Adenovirus E1A recruits the human Paf1 complex to enhance transcriptional elongation
- PMID: 24600005
- PMCID: PMC4019120
- DOI: 10.1128/JVI.03518-13
Adenovirus E1A recruits the human Paf1 complex to enhance transcriptional elongation
Abstract
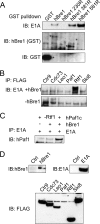
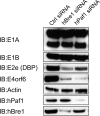
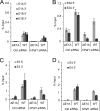
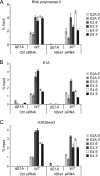
During infection by human adenovirus (HAdV), the proteins encoded by the early region 1A (E1A) gene bind and appropriate components of the cellular transcriptional machinery to activate the transcription of viral early genes. Previously, we identified roles for the human Bre1 (hBre1) and hPaf1 complexes in E1A-mediated transcriptional activation of HAdV early genes. Here we show that E1A binds hBre1 directly and that this complex targets the hPaf1 complex via the Rtf1 subunit. Depletion of hPaf1 reduces E1A-dependent activation of transcription from the E2e, E3, and E4 viral transcription units, and this does not result from a reduced ability of RNA polymerase II to be recruited to the promoter-proximal regions of these genes. In contrast, depletion of hPaf1 reduces the occupancy of RNA polymerase II across these transcription units. This is accompanied by reductions in the level of H3K36 trimethylation, a posttranslational histone modification associated with efficient transcriptional elongation, and the number of full-length transcripts from these genes. Together, these results indicate that E1A uses hBre1 to recruit the hPaf1 complex in order to optimally activate viral early transcription by enhancing transcriptional elongation.
Importance: This work provides the mechanism by which the hPaf1 complex contributes to E1A-dependent activation of early gene transcription. The work also demonstrates that E1A induces gene expression by stimulating transcriptional elongation, in addition to its better-characterized effects on transcriptional initiation.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

