Comparison of varicella-zoster virus RNA sequences in human neurons and fibroblasts
- PMID: 24600007
- PMCID: PMC4019124
- DOI: 10.1128/JVI.00476-14
Comparison of varicella-zoster virus RNA sequences in human neurons and fibroblasts
Abstract
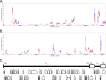
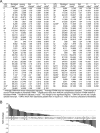
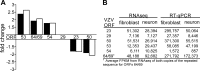
Varicella-zoster virus (VZV) infection causes varicella, after which the virus becomes latent in ganglionic neurons. In tissue culture, VZV-infected human neurons remain viable at 2 weeks, whereas fibroblasts develop cytopathology. Next-generation RNA sequencing was used to compare VZV transcriptomes in neurons and fibroblasts and identified only 12 differentially transcribed genes of the 70 annotated VZV open reading frames (ORFs), suggesting that defective virus transcription does not account for the lack of cell death in VZV-infected neurons in vitro.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

