Machine learning for neuroimaging with scikit-learn
- PMID: 24600388
- PMCID: PMC3930868
- DOI: 10.3389/fninf.2014.00014
Machine learning for neuroimaging with scikit-learn
Abstract
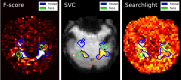
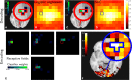
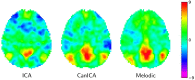
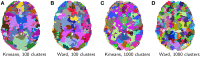
Statistical machine learning methods are increasingly used for neuroimaging data analysis. Their main virtue is their ability to model high-dimensional datasets, e.g., multivariate analysis of activation images or resting-state time series. Supervised learning is typically used in decoding or encoding settings to relate brain images to behavioral or clinical observations, while unsupervised learning can uncover hidden structures in sets of images (e.g., resting state functional MRI) or find sub-populations in large cohorts. By considering different functional neuroimaging applications, we illustrate how scikit-learn, a Python machine learning library, can be used to perform some key analysis steps. Scikit-learn contains a very large set of statistical learning algorithms, both supervised and unsupervised, and its application to neuroimaging data provides a versatile tool to study the brain.
Keywords: Python; machine learning; neuroimaging; scikit-learn; statistical learning.
Figures

References
-
- Detre G., Polyn S., Moore C., Natu V., Singer B., Cohen J., et al. (2006). The multi-voxel pattern analysis (mvpa) toolbox, in Poster Presented at the Annual Meeting of the Organization for Human Brain Mapping (Florence, Italy: ). Available online at: http://www.csbmb.princeton.edu/mvpa
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

