Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants
- PMID: 24600444
- PMCID: PMC3928675
- DOI: 10.3389/fmicb.2014.00064
Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants
Abstract
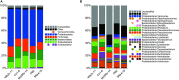
Plant-associated bacteria fulfill important functions for plant growth and health. However, our knowledge about the impact of bacterial treatments on the host's microbiome and physiology is limited. The present study was conducted to assess the impact of bacterial inoculants on the microbiome of chamomile plants Chamomilla recutita (L.) Rauschert grown in a field under organic management in Egypt. Chamomile seedlings were inoculated with three indigenous Gram-positive strains (Streptomyces subrutilus Wbn2-11, Bacillus subtilis Co1-6, Paenibacillus polymyxa Mc5Re-14) from Egypt and three European Gram-negative strains (Pseudomonas fluorescens L13-6-12, Stenotrophomonas rhizophila P69, Serratia plymuthica 3Re4-18) already known for their beneficial plant-microbe interaction. Molecular fingerprints of 16S rRNA gene as well as real-time PCR analyses did not show statistically significant differences for all applied bacterial antagonists compared to the control. In contrast, a pyrosequencing analysis of the 16S rRNA gene libraries revealed significant differences in the community structure of bacteria between the treatments. These differences could be clearly shown by a shift within the community structure and corresponding beta-diversity indices. Moreover, B. subtilis Co1-6 and P. polymyxa Mc5Re-14 showed an enhancement of the bioactive secondary metabolite apigenin-7-O-glucoside. This indicates a possible new function of bacterial inoculants: to interact with the plant microbiome as well as to influence the plant metabolome.
Keywords: bioactive secondary metabolites; biological control agents; chamomile; microbial communities; soil-borne pathogens.
Figures






References
-
- Arkhipova T. N., Prinsen E., Veselov S. U., Martinenko E. V., Melentiev A. I., Kudoyarova G. R. (2007). Cytokinin producing bacteria enhance plant growth in drying soil. Plant Soil 292, 305–315 10.1007/s11104-007-9233-5 - DOI
-
- Bauer R., Wagner H. (1991). Äußere Einflüsse auf den Flavonoidgehalt von Kamillenblüten. Sci. Pharm. 59, 3
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

