Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals
- PMID: 24603613
- PMCID: PMC3945117
- DOI: 10.1371/journal.pgen.1004155
Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals
Abstract
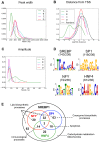
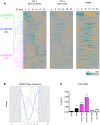
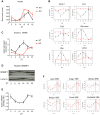
In mammals, the circadian clock allows them to anticipate and adapt physiology around the 24 hours. Conversely, metabolism and food consumption regulate the internal clock, pointing the existence of an intricate relationship between nutrient state and circadian homeostasis that is far from being understood. The Sterol Regulatory Element Binding Protein 1 (SREBP1) is a key regulator of lipid homeostasis. Hepatic SREBP1 function is influenced by the nutrient-response cycle, but also by the circadian machinery. To systematically understand how the interplay of circadian clock and nutrient-driven rhythm regulates SREBP1 activity, we evaluated the genome-wide binding of SREBP1 to its targets throughout the day in C57BL/6 mice. The recruitment of SREBP1 to the DNA showed a highly circadian behaviour, with a maximum during the fed status. However, the temporal expression of SREBP1 targets was not always synchronized with its binding pattern. In particular, different expression phases were observed for SREBP1 target genes depending on their function, suggesting the involvement of other transcription factors in their regulation. Binding sites for Hepatocyte Nuclear Factor 4 (HNF4) were specifically enriched in the close proximity of SREBP1 peaks of genes, whose expression was shifted by about 8 hours with respect to SREBP1 binding. Thus, the cross-talk between hepatic HNF4 and SREBP1 may underlie the expression timing of this subgroup of SREBP1 targets. Interestingly, the proper temporal expression profile of these genes was dramatically changed in Bmal1-/- mice upon time-restricted feeding, for which a rhythmic, but slightly delayed, binding of SREBP1 was maintained. Collectively, our results show that besides the nutrient-driven regulation of SREBP1 nuclear translocation, a second layer of modulation of SREBP1 transcriptional activity, strongly dependent from the circadian clock, exists. This system allows us to fine tune the expression timing of SREBP1 target genes, thus helping to temporally separate the different physiological processes in which these genes are involved.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Moynihan Ramsey K, Marcheva B, Kohsaka A, Bass J (2007) The Clockwork of Metabolism. Annual Review of Nutrition 27: 219–240. - PubMed
-
- Stokkan K-A, Yamazaki S, Tei H, Sakaki Y, Menaker M (2001) Entrainment of the Circadian Clock in the Liver by Feeding. Science 291: 490–493. - PubMed
-
- Di Lorenzo L, De Pergola G, Zocchetti C, L'abbate N, Basso A, et al. (2003) Effect of shift work on body mass index: results of a study performed in 319 glucose-tolerant men working in a Southern Italian industry. Int J Obes Relat Metab Disord 27: 1353–1358. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

