GC-rich DNA elements enable replication origin activity in the methylotrophic yeast Pichia pastoris
- PMID: 24603708
- PMCID: PMC3945215
- DOI: 10.1371/journal.pgen.1004169
GC-rich DNA elements enable replication origin activity in the methylotrophic yeast Pichia pastoris
Abstract
The well-studied DNA replication origins of the model budding and fission yeasts are A/T-rich elements. However, unlike their yeast counterparts, both plant and metazoan origins are G/C-rich and are associated with transcription start sites. Here we show that an industrially important methylotrophic budding yeast, Pichia pastoris, simultaneously employs at least two types of replication origins--a G/C-rich type associated with transcription start sites and an A/T-rich type more reminiscent of typical budding and fission yeast origins. We used a suite of massively parallel sequencing tools to map and dissect P. pastoris origins comprehensively, to measure their replication dynamics, and to assay the global positioning of nucleosomes across the genome. Our results suggest that some functional overlap exists between promoter sequences and G/C-rich replication origins in P. pastoris and imply an evolutionary bifurcation of the modes of replication initiation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
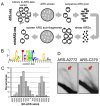



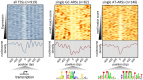

References
-
- Méchali M (2010) Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738 doi:10.1038/nrm2976 - DOI - PubMed
-
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, et al. (2011) Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174 doi:10.1101/gr.116038.110 - DOI - PMC - PubMed
-
- Dellino GI, Cittaro D, Piccioni R, Luzi L, Banfi S, et al. (2013) Genome-wide mapping of human DNA-replication origins: Levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res 23: 1–11 doi:10.1101/gr.142331.112 - DOI - PMC - PubMed
-
- Costas C, la Paz Sanchez de M, Stroud H, Yu Y, Oliveros JC, et al. (2011) Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat Struct Mol Biol 18: 395–400 doi:10.1038/nsmb.1988 - DOI - PMC - PubMed
-
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, et al. (2010) Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci USA 107: 139–144 doi:10.1073/pnas.0912402107 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

