The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli
- PMID: 24603758
- PMCID: PMC3946193
- DOI: 10.1371/journal.pone.0090447
The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli
Erratum in
- PLoS One. 2014;9(6):e100908
Abstract
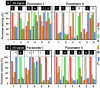
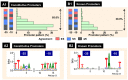
The promoter selectivity of Escherichia coli RNA polymerase is determined by the sigma subunit with promoter recognition activity. The model prokaryote Escherichia coli contains seven species of the sigma subunit, each recognizing a specific set of promoters. The major sigma subunit, sigma-70 encoded by rpoD, plays a major role in transcription of growth-related genes. Concomitant with the increase in detection of promoters functioning in vivo under various stressful conditions, the variation is expanding in the consensus sequence of RpoD promoters. In order to identify the canonical sequence of "constitutive promoters" that are recognized by the RNA polymerase holoenzyme containing RpoD sigma in the absence of supporting transcription factors, an in vitro mixed transcription assay was carried out using a whole set of variant promoters, each harboring one base replacement, within the model promoter with the conserved -35 and -10 sequences of RpoD promoters. The consensus sequences, TTGACA(-35) and TATAAT(-10), were identified to be ideal for the maximum level of open complex formation and the highest rate of promoter opening, respectively. For identification of the full range of constitutive promoters on the E. coli genome, a total of 2,701 RpoD holoenzyme-binding sites were identified by Genomic SELEX screening, and using the reconfirmed consensus promoter sequence, a total of maximum 669 constitutive promoters were identified, implying that the majority of hitherto identified promoters represents the TF-dependent "inducible promoters". One unique feature of the constitutive promoters is the high level of promoter sequence conservation, about 85% carrying five-out-of-six agreements with -35 or -10 consensus sequence. The list of constitutive promoters provides the community resource toward estimation of the inducible promoters that operate under various stressful conditions in nature.
Conflict of interest statement
Figures



References
-
- Gruber TM, Gross CA (2003) Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57: 441–466. - PubMed
-
- Ishihama A (2000) Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol 54: 499–518. - PubMed
-
- Ishihama A (2010) Prokaryotic genome regulation: Multi-factor promoters, multi-target regulators and hierarchic networks. FEMS Microbial Rev 34: 628–645. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

