Genetic dissection of Anopheles gambiae gut epithelial responses to Serratia marcescens
- PMID: 24603764
- PMCID: PMC3946313
- DOI: 10.1371/journal.ppat.1003897
Genetic dissection of Anopheles gambiae gut epithelial responses to Serratia marcescens
Abstract
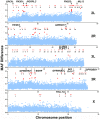
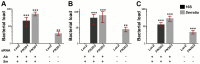
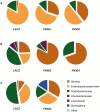
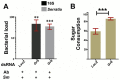
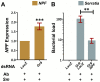
Genetic variation in the mosquito Anopheles gambiae profoundly influences its ability to transmit malaria. Mosquito gut bacteria are shown to influence the outcome of infections with Plasmodium parasites and are also thought to exert a strong drive on genetic variation through natural selection; however, a link between antibacterial effects and genetic variation is yet to emerge. Here, we combined SNP genotyping and expression profiling with phenotypic analyses of candidate genes by RNAi-mediated silencing and 454 pyrosequencing to investigate this intricate biological system. We identified 138 An. gambiae genes to be genetically associated with the outcome of Serratia marcescens infection, including the peptidoglycan recognition receptor PGRPLC that triggers activation of the antibacterial IMD/REL2 pathway and the epidermal growth factor receptor EGFR. Silencing of three genes encoding type III fibronectin domain proteins (FN3Ds) increased the Serratia load and altered the gut microbiota composition in favor of Enterobacteriaceae. These data suggest that natural genetic variation in immune-related genes can shape the bacterial population structure of the mosquito gut with high specificity. Importantly, FN3D2 encodes a homolog of the hypervariable pattern recognition receptor Dscam, suggesting that pathogen-specific recognition may involve a broader family of immune factors. Additionally, we showed that silencing the gene encoding the gustatory receptor Gr9 that is also associated with the Serratia infection phenotype drastically increased Serratia levels. The Gr9 antibacterial activity appears to be related to mosquito feeding behavior and to mostly rely on changes of neuropeptide F expression, together suggesting a behavioral immune response following Serratia infection. Our findings reveal that the mosquito response to oral Serratia infection comprises both an epithelial and a behavioral immune component.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Riehle MM, Markianos K, Niare O, Xu J, Li J, et al. (2006) Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science 312: 577–579. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/E002641/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G1100339/MRC_/Medical Research Council/United Kingdom
- BB/K009338/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 086723/Z/08/A/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

