Mechanisms underlying differential response to estrogen-induced apoptosis in long-term estrogen-deprived breast cancer cells
- PMID: 24604139
- PMCID: PMC4144033
- DOI: 10.3892/ijo.2014.2329
Mechanisms underlying differential response to estrogen-induced apoptosis in long-term estrogen-deprived breast cancer cells
Abstract
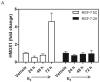
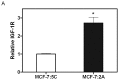
Models of long-term estrogen-deprived breast cancer cells are utilized in the laboratory to mimic clinical aromatase inhibitor-resistant breast cancer and serve as a tool to discover new therapeutic strategies. The MCF-7:5C and MCF-7:2A subclones were generated through long-term estrogen deprivation of estrogen receptor (ER)-positive MCF-7 cells, and represent anti-hormone‑resistant breast cancer. MCF-7:5C cells paradoxically undergo estrogen-induced apoptosis within seven days of estrogen (estradiol, E2) treatment; MCF-7:2A cells also experience E(2)-induced apoptosis but evade dramatic cell death until approximately 14 days of treatment. To discover and define the mechanisms by which MCF-7:2A cells survive two weeks of E(2) treatment, systematic experiments were performed in this study. The data suggest that MCF-7:2A cells employ stronger antioxidant defense mechanisms than do MCF-7:5C cells, and that oxidative stress is ultimately required for MCF-7:2A cells to die in response to E2 treatment. Tumor necrosis factor (TNF) family member activation is also essential for E(2)-induced apoptosis to occur in MCF-7:2A cells; upregulation of TNFα occurs simultaneously with oxidative stress activation. Although the unfolded protein response (UPR) signaling pattern is similar to that in MCF-7:5C cells, it is not sufficient to cause cell death in MCF-7:2A cells. Additionally, increased insulin-like growth factor receptor β (IGF-1Rβ) confers a mechanism of growth and anti-apoptotic advantage in MCF-7:2A cells.
Figures












Similar articles
-
Targeting Peroxisome Proliferator-Activated Receptor γ to Increase Estrogen-Induced Apoptosis in Estrogen-Deprived Breast Cancer Cells.Mol Cancer Ther. 2018 Dec;17(12):2732-2745. doi: 10.1158/1535-7163.MCT-18-0088. Epub 2018 Sep 17. Mol Cancer Ther. 2018. PMID: 30224430 Free PMC article.
-
Buthionine sulfoximine sensitizes antihormone-resistant human breast cancer cells to estrogen-induced apoptosis.Breast Cancer Res. 2008;10(6):R104. doi: 10.1186/bcr2208. Epub 2008 Dec 5. Breast Cancer Res. 2008. PMID: 19061505 Free PMC article.
-
Inhibition of c-Src blocks oestrogen-induced apoptosis and restores oestrogen-stimulated growth in long-term oestrogen-deprived breast cancer cells.Eur J Cancer. 2014 Jan;50(2):457-68. doi: 10.1016/j.ejca.2013.10.001. Epub 2013 Oct 30. Eur J Cancer. 2014. PMID: 24183378 Free PMC article.
-
Selective estrogen-induced apoptosis in breast cancer.Steroids. 2014 Nov;90:60-70. doi: 10.1016/j.steroids.2014.06.003. Epub 2014 Jun 11. Steroids. 2014. PMID: 24929046 Review.
-
The new biology of estrogen-induced apoptosis applied to treat and prevent breast cancer.Endocr Relat Cancer. 2015 Feb;22(1):R1-31. doi: 10.1530/ERC-14-0448. Epub 2014 Oct 22. Endocr Relat Cancer. 2015. PMID: 25339261 Free PMC article. Review.
Cited by
-
PERK, Beyond an Unfolded Protein Response Sensor in Estrogen-Induced Apoptosis in Endocrine-Resistant Breast Cancer.Mol Cancer Res. 2022 Feb;20(2):193-201. doi: 10.1158/1541-7786.MCR-21-0702. Epub 2021 Nov 2. Mol Cancer Res. 2022. PMID: 34728551 Free PMC article. Review.
-
Acquired resistance to selective estrogen receptor modulators (SERMs) in clinical practice (tamoxifen & raloxifene) by selection pressure in breast cancer cell populations.Steroids. 2014 Nov;90:44-52. doi: 10.1016/j.steroids.2014.06.002. Epub 2014 Jun 12. Steroids. 2014. PMID: 24930824 Free PMC article.
-
Estrogen Receptor and the Unfolded Protein Response: Double-Edged Swords in Therapy for Estrogen Receptor-Positive Breast Cancer.Target Oncol. 2022 Mar;17(2):111-124. doi: 10.1007/s11523-022-00870-5. Epub 2022 Mar 15. Target Oncol. 2022. PMID: 35290592 Free PMC article. Review.
-
Estrogen Receptor Complex to Trigger or Delay Estrogen-Induced Apoptosis in Long-Term Estrogen Deprived Breast Cancer.Front Endocrinol (Lausanne). 2022 Mar 10;13:869562. doi: 10.3389/fendo.2022.869562. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35360069 Free PMC article. Review.
-
Targeting Peroxisome Proliferator-Activated Receptor γ to Increase Estrogen-Induced Apoptosis in Estrogen-Deprived Breast Cancer Cells.Mol Cancer Ther. 2018 Dec;17(12):2732-2745. doi: 10.1158/1535-7163.MCT-18-0088. Epub 2018 Sep 17. Mol Cancer Ther. 2018. PMID: 30224430 Free PMC article.
References
-
- Jiang SY, Wolf DM, Yingling JM, Chang C, Jordan VC. An estrogen receptor positive MCF-7 clone that is resistant to anti-estrogens and estradiol. Mol Cell Endocrinol. 1992;90:77–86. - PubMed
-
- Lewis JS, Osipo C, Meeke K, Jordan VC. Estrogen-induced apoptosis in a breast cancer model resistant to long-term estrogen withdrawal. J Steroid Biochem Mol Biol. 2005;94:131–141. - PubMed
-
- Pink JJ, Jiang SY, Fritsch M, Jordan VC. An estrogen-independent MCF-7 breast cancer cell line which contains a novel 80-kilodalton estrogen receptor-related protein. Cancer Res. 1995;55:2583–2590. - PubMed
-
- Pink JJ, Jordan VC. Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res. 1996;56:2321–2330. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

