Epigenetic estimation of age in humpback whales
- PMID: 24606053
- PMCID: PMC4314680
- DOI: 10.1111/1755-0998.12247
Epigenetic estimation of age in humpback whales
Abstract
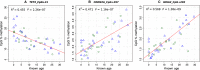
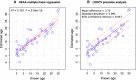
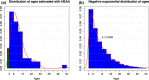
Age is a fundamental aspect of animal ecology, but is difficult to determine in many species. Humpback whales exemplify this as they have a lifespan comparable to humans, mature sexually as early as 4 years and have no reliable visual age indicators after their first year. Current methods for estimating humpback age cannot be applied to all individuals and populations. Assays for human age have recently been developed based on age-induced changes in DNA methylation of specific genes. We used information on age-associated DNA methylation in human and mouse genes to identify homologous gene regions in humpbacks. Humpback skin samples were obtained from individuals with a known year of birth and employed to calibrate relationships between cytosine methylation and age. Seven of 37 cytosines assayed for methylation level in humpback skin had significant age-related profiles. The three most age-informative cytosine markers were selected for a humpback epigenetic age assay. The assay has an R(2) of 0.787 (P = 3.04e-16) and predicts age from skin samples with a standard deviation of 2.991 years. The epigenetic method correctly determined which of parent-offspring pairs is the parent in more than 93% of cases. To demonstrate the potential of this technique, we constructed the first modern age profile of humpback whales off eastern Australia and compared the results to population structure 5 decades earlier. This is the first epigenetic age estimation method for a wild animal species and the approach we took for developing it can be applied to many other nonmodel organisms.
Keywords: DNA methylation; age; cetacean; epigenetics; population.
© 2014 The Authors. Molecular Ecology Resources Published by John Wiley & Sons Ltd.
Figures

References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. - PubMed
-
- Beverton RJH, Holt SJ. A review of methods for estimating mortality rates in exploited fish populations, with special reference to sources of bias in catch sampling. Rapp. P.-V. Réun. Ciem. 1956;140:67–83.
-
- Baker CS, Clapham PJ. Modelling the past and future of whales and whaling. Trends in Ecology & Evolution. 2004;19:365–371. - PubMed
-
- Barlow J, Clapham PJ. A new birth-interval approach to estimating demographic parameters of humpback whales. Ecology. 1997;78:535–546.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

