Demystifying heparan sulfate-protein interactions
- PMID: 24606135
- PMCID: PMC7851832
- DOI: 10.1146/annurev-biochem-060713-035314
Demystifying heparan sulfate-protein interactions
Abstract
Numerous proteins, including cytokines and chemokines, enzymes and enzyme inhibitors, extracellular matrix proteins, and membrane receptors, bind heparin. Although they are traditionally classified as heparin-binding proteins, under normal physiological conditions these proteins actually interact with the heparan sulfate chains of one or more membrane or extracellular proteoglycans. Thus, they are more appropriately classified as heparan sulfate-binding proteins (HSBPs). This review provides an overview of the various modes of interaction between heparan sulfate and HSBPs, emphasizing biochemical and structural insights that improve our understanding of the many biological functions of heparan sulfate.
Keywords: glycan–protein interaction; glycosaminoglycan; heparan sulfate–binding domain; heparin-binding protein; oligomerization; proteoglycan.
Figures
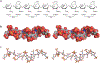



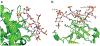
References
-
- Guerardel Y, Czeszak X, Sumanovski LT, Karamanos Y, Popescu O, et al. 2004. Molecular fingerprinting of carbohydrate structure phenotypes of three Porifera proteoglycan-like glyconectins. J. Biol. Chem 279:15591–603 - PubMed
-
- Yamada S, Morimoto H, Fujisawa T, Sugahara K. 2007. Glycosaminoglycans in Hydra magnipapillata (Hydrozoa, Cnidaria): demonstration of chondroitin in the developing nematocyst, sting organelle, and structural characterization of glycosaminoglycans. Glycobiology 17:886–94 - PubMed
-
- Bishop JR, Schuksz M, Esko JD. 2007. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446:1030–37 - PubMed

