S-adenosylmethionine levels regulate the schwann cell DNA methylome
- PMID: 24607226
- PMCID: PMC3960855
- DOI: 10.1016/j.neuron.2014.01.037
S-adenosylmethionine levels regulate the schwann cell DNA methylome
Abstract
Axonal myelination is essential for rapid saltatory impulse conduction in the nervous system, and malformation or destruction of myelin sheaths leads to motor and sensory disabilities. DNA methylation is an essential epigenetic modification during mammalian development, yet its role in myelination remains obscure. Here, using high-resolution methylome maps, we show that DNA methylation could play a key gene regulatory role in peripheral nerve myelination and that S-adenosylmethionine (SAMe), the principal methyl donor in cytosine methylation, regulates the methylome dynamics during this process. Our studies also point to a possible role of SAMe in establishing the aberrant DNA methylation patterns in a mouse model of diabetic neuropathy, implicating SAMe in the pathogenesis of this disease. These critical observations establish a link between SAMe and DNA methylation status in a defined biological system, providing a mechanism that could direct methylation changes during cellular differentiation and in diverse pathological situations.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures






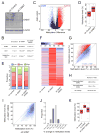
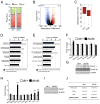
References
-
- Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–719. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

