The EMPRES-i genetic module: a novel tool linking epidemiological outbreak information and genetic characteristics of influenza viruses
- PMID: 24608033
- PMCID: PMC3945526
- DOI: 10.1093/database/bau008
The EMPRES-i genetic module: a novel tool linking epidemiological outbreak information and genetic characteristics of influenza viruses
Abstract
Combining epidemiological information, genetic characterization and geomapping in the analysis of influenza can contribute to a better understanding and description of influenza epidemiology and ecology, including possible virus reassortment events. Furthermore, integration of information such as agroecological farming system characteristics can provide new knowledge on risk factors of influenza emergence and spread. Integrating viral characteristics into an animal disease information system is therefore expected to provide a unique tool to trace-and-track particular virus strains; generate clade distributions and spatiotemporal clusters; screen for distribution of viruses with specific molecular markers; identify potential risk factors; and analyze or map viral characteristics related to vaccines used for control and/or prevention. For this purpose, a genetic module was developed within EMPRES-i (FAO's global animal disease information system) linking epidemiological information from influenza events with virus characteristics and enabling combined analysis. An algorithm was developed to act as the interface between EMPRES-i disease event data and publicly available influenza virus sequences in OpenfluDB. This algorithm automatically computes potential links between outbreak event and sequences, which are subsequently manually validated by experts. Subsequently, other virus characteristics such as antiviral resistance can then be associated to outbreak data. To visualize such characteristics on a geographic map, shape files with virus characteristics to overlay on other EMPRES-i map layers (e.g. animal densities) can be generated. The genetic module allows export of associated epidemiological and sequence data for further analysis. FAO has made this tool available for scientists and policy makers. Contributions are expected from users to improve and validate the number of linked influenza events and isolate information as well as the quality of information. Possibilities to interconnect with other influenza sequence databases or to expand the genetic module to other viral diseases (e.g. foot and mouth disease) are being explored. Database OpenfluDB URL: http://openflu.vital-it.ch Database EMPRES-i URL: http://EMPRES-i.fao.org/.
Figures

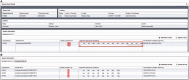
 or individual links under ‘Segments Sequences’ for GenBank will open a new browser window or link to display the sequence record.
or individual links under ‘Segments Sequences’ for GenBank will open a new browser window or link to display the sequence record.
References
-
- Muellner P, Zadoks RN, Perez AM, et al. French. The integration of molecular tools into veterinary and spatial epidemiology. Spat. Spatiotemporal Epidemiol. 2011;2:159–171. - PubMed
-
- Archie EA, Luikart G, Ezenwa VO. Infecting epidemiology with genetics: a new frontier in disease ecology. Trends Ecol. Evol. 2009;24:21–30. - PubMed
-
- Zambon MC. Epidemiology and pathogenesis of influenza. J. Antimicrob. Chemother. 1999;44:3–9. - PubMed
-
- Lam TT-Y, Hon CC, Tang JW. Use of phylogenetics in the molecular epidemiology and evolutionary studies of viral infections. Crit. Rev. Clin. Lab. Sci. 2010;47:5–49. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

