Variation in RNA-Seq transcriptome profiles of peripheral whole blood from healthy individuals with and without globin depletion
- PMID: 24608128
- PMCID: PMC3946641
- DOI: 10.1371/journal.pone.0091041
Variation in RNA-Seq transcriptome profiles of peripheral whole blood from healthy individuals with and without globin depletion
Abstract
Background: The molecular profile of circulating blood can reflect physiological and pathological events occurring in other tissues and organs of the body and delivers a comprehensive view of the status of the immune system. Blood has been useful in studying the pathobiology of many diseases. It is accessible and easily collected making it ideally suited to the development of diagnostic biomarker tests. The blood transcriptome has a high complement of globin RNA that could potentially saturate next-generation sequencing platforms, masking lower abundance transcripts. Methods to deplete globin mRNA are available, but their effect has not been comprehensively studied in peripheral whole blood RNA-Seq data. In this study we aimed to assess technical variability associated with globin depletion in addition to assessing general technical variability in RNA-Seq from whole blood derived samples.
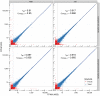
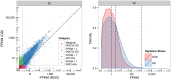
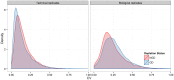
Results: We compared technical and biological replicates having undergone globin depletion or not and found that the experimental globin depletion protocol employed removed approximately 80% of globin transcripts, improved the correlation of technical replicates, allowed for reliable detection of thousands of additional transcripts and generally increased transcript abundance measures. Differential expression analysis revealed thousands of genes significantly up-regulated as a result of globin depletion. In addition, globin depletion resulted in the down-regulation of genes involved in both iron and zinc metal ion bonding.
Conclusions: Globin depletion appears to meaningfully improve the quality of peripheral whole blood RNA-Seq data, and may improve our ability to detect true biological variation. Some concerns remain, however. Key amongst them the significant reduction in RNA yields following globin depletion. More generally, our investigation of technical and biological variation with and without globin depletion finds that high-throughput sequencing by RNA-Seq is highly reproducible within a large dynamic range of detection and provides an accurate estimation of RNA concentration in peripheral whole blood. High-throughput sequencing is thus a promising technology for whole blood transcriptomics and biomarker discovery.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

