Mdmx promotes genomic instability independent of p53 and Mdm2
- PMID: 24608433
- PMCID: PMC4160436
- DOI: 10.1038/onc.2014.27
Mdmx promotes genomic instability independent of p53 and Mdm2
Abstract
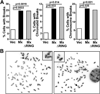
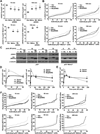
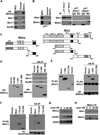
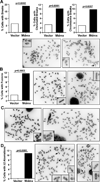
The oncogene Mdmx is overexpressed in many human malignancies, and together with Mdm2, negatively regulates the p53 tumor suppressor. However, a p53-independent function of Mdmx that impacts genome stability has been described, but this function is not well understood. In the present study, we determined that of the 13 different cancer types evaluated, 6-90% of those that had elevated levels of Mdmx had concurrent inactivation (mutated or deleted) of p53. We show elevated levels of Mdmx-inhibited double-strand DNA break repair and induced chromosome and chromatid breaks independent of p53, leading to genome instability. Mdmx impaired early DNA damage-response signaling, such as phosphorylation of the serine/threonine-glutamine motif, mediated by the ATM kinase. Moreover, we identified Mdmx associated with Nbs1 of the Mre11-Rad50-Nbs1 (MRN) DNA repair complex, and this association increased upon DNA damage and was detected at chromatin. Elevated Mdmx levels also increased cellular transformation in a p53-independent manner. Unexpectedly, all Mdmx-mediated phenotypes also occurred in cells lacking Mdm2 and were independent of the Mdm2-binding domain (RING) of Mdmx. Therefore, Mdmx-mediated inhibition of the DNA damage response resulted in delayed DNA repair and increased genome instability and transformation independent of p53 and Mdm2. Our results reveal a novel p53- and Mdm2-independent oncogenic function of Mdmx that provides new insight into the many cancers that overexpress Mdmx.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures

References
-
- Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27–41. - PubMed
-
- Lundgren K, Montes de Oca Luna R, McNeill YB, Emerick EP, Spencer B, Barfield CR, et al. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 1997;11:714–725. - PubMed
-
- Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem. 2005;280:18771–18781. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

