SUMOylation of Myc-family proteins
- PMID: 24608896
- PMCID: PMC3946688
- DOI: 10.1371/journal.pone.0091072
SUMOylation of Myc-family proteins
Abstract
Myc-family proteins are key controllers of the metabolic and proliferative status of the cell, and are subjected to a complex network of regulatory events that guarantee their efficient and fast modulation by extracellular stimuli. Hence, unbalances in regulatory mechanisms leading to altered Myc levels or activities are often reported in cancer cells. Here we show that c- and N-Myc are conjugated to SUMO proteins at conserved lysines in their C-terminal domain. No obvious effects of SUMOylation were detected on bulk N-Myc stability or activities, including the regulation of transcription, proliferation or apoptosis. N-Myc SUMOylation could be induced by cellular stresses, such as heat shock and proteasome inhibition, and in all instances concerned a small fraction of the N-Myc protein. We surmise that, as shown for other substrates, SUMOylation may be part of a quality-control mechanism acting on misfolded Myc proteins.
Conflict of interest statement
Figures
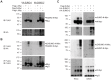
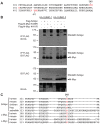
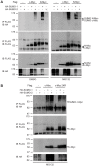
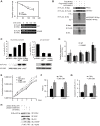

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

