High Rhodotorula sequences in skin transcriptome of patients with diffuse systemic sclerosis
- PMID: 24608988
- PMCID: PMC4102619
- DOI: 10.1038/jid.2014.127
High Rhodotorula sequences in skin transcriptome of patients with diffuse systemic sclerosis
Abstract
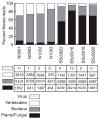
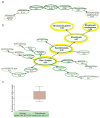
Previous studies have suggested a role for pathogens as a trigger of systemic sclerosis (SSc), although neither a pathogen nor a mechanism of pathogenesis is known. Here we show enrichment of Rhodotorula sequences in the skin of patients with early, diffuse SSc compared with that in normal controls. RNA-seq was performed on four SSc patients and four controls, to a depth of 200 million reads per patient. Data were analyzed to quantify the nonhuman sequence reads in each sample. We found little difference between bacterial microbiome and viral read counts, but found a significant difference between the read counts for a mycobiome component, R. glutinis. Normal samples contained almost no detected R. glutinis or other Rhodotorula sequence reads (mean score 0.021 for R. glutinis, 0.024 for all Rhodotorula). In contrast, SSc samples had a mean score of 5.039 for R. glutinis (5.232 for Rhodotorula). We were able to assemble the D1-D2 hypervariable region of the 28S ribosomal RNA (rRNA) of R. glutinis from each of the SSc samples. Taken together, these results suggest that R. glutinis may be present in the skin of early SSc patients at higher levels than in normal skin, raising the possibility that it may be triggering the inflammatory response found in SSc.
Conflict of interest statement
Dr. Whitfield has filed patents for gene expression biomarkers in systemic sclerosis and is a scientific founder of Celdara Medical LLC. The remaining authors declare no conflict of interest.
Figures



References
-
- Alvarez-Fernández JA, Quirce S, Calleja JL, et al. Hypersensitivity pneumonitis due to an ultrasonic humidifier. Allergy. 1998;53:210–2. - PubMed
-
- Biswas SK, Yokoyama K, Nishimura K, et al. Molecular phylogenetics of the genus Rhodotorula and related basidiomycetous yeasts inferred from the mitochondrial cytochrome b gene. Int J Syst Evol Microbiol. 2001;51:1191–9. - PubMed
-
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the nonspecialist. Nucleic Acids Res. 2008;36:W465–469. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

