Glycoprotein folding and quality-control mechanisms in protein-folding diseases
- PMID: 24609034
- PMCID: PMC3944493
- DOI: 10.1242/dmm.014589
Glycoprotein folding and quality-control mechanisms in protein-folding diseases
Abstract
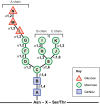
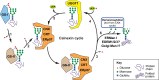
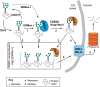
Biosynthesis of proteins--from translation to folding to export--encompasses a complex set of events that are exquisitely regulated and scrutinized to ensure the functional quality of the end products. Cells have evolved to capitalize on multiple post-translational modifications in addition to primary structure to indicate the folding status of nascent polypeptides to the chaperones and other proteins that assist in their folding and export. These modifications can also, in the case of irreversibly misfolded candidates, signal the need for dislocation and degradation. The current Review focuses on the glycoprotein quality-control (GQC) system that utilizes protein N-glycosylation and N-glycan trimming to direct nascent glycopolypeptides through the folding, export and dislocation pathways in the endoplasmic reticulum (ER). A diverse set of pathological conditions rooted in defective as well as over-vigilant ER quality-control systems have been identified, underlining its importance in human health and disease. We describe the GQC pathways and highlight disease and animal models that have been instrumental in clarifying our current understanding of these processes.
Keywords: ER export; ER quality control; ER-associated degradation; Glycoprotein folding; N-glycosylation.
Figures

References
-
- Aebi M., Bernasconi R., Clerc S., Molinari M. (2010). N-glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 35, 74–82 - PubMed
-
- Appenzeller C., Andersson H., Kappeler F., Hauri H. P. (1999). The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat. Cell Biol. 1, 330–334 - PubMed
-
- Appenzeller-Herzog C., Roche A. C., Nufer O., Hauri H. P. (2004). pH-induced conversion of the transport lectin ERGIC-53 triggers glycoprotein release. J. Biol. Chem. 279, 12943–12950 - PubMed
-
- Apweiler R., Hermjakob H., Sharon N. (1999). On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

