The effect of linkage on establishment and survival of locally beneficial mutations
- PMID: 24610861
- PMCID: PMC4012489
- DOI: 10.1534/genetics.114.163477
The effect of linkage on establishment and survival of locally beneficial mutations
Abstract
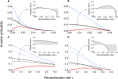
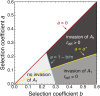
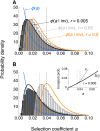
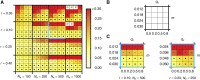
We study invasion and survival of weakly beneficial mutations arising in linkage to an established migration-selection polymorphism. Our focus is on a continent-island model of migration, with selection at two biallelic loci for adaptation to the island environment. Combining branching and diffusion processes, we provide the theoretical basis for understanding the evolution of islands of divergence, the genetic architecture of locally adaptive traits, and the importance of so-called "divergence hitchhiking" relative to other mechanisms, such as "genomic hitchhiking", chromosomal inversions, or translocations. We derive approximations to the invasion probability and the extinction time of a de novo mutation. Interestingly, the invasion probability is maximized at a nonzero recombination rate if the focal mutation is sufficiently beneficial. If a proportion of migrants carries a beneficial background allele, the mutation is less likely to become established. Linked selection may increase the survival time by several orders of magnitude. By altering the timescale of stochastic loss, it can therefore affect the dynamics at the focal site to an extent that is of evolutionary importance, especially in small populations. We derive an effective migration rate experienced by the weakly beneficial mutation, which accounts for the reduction in gene flow imposed by linked selection. Using the concept of the effective migration rate, we also quantify the long-term effects on neutral variation embedded in a genome with arbitrarily many sites under selection. Patterns of neutral diversity change qualitatively and quantitatively as the position of the neutral locus is moved along the chromosome. This will be useful for population-genomic inference. Our results strengthen the emerging view that physically linked selection is biologically relevant if linkage is tight or if selection at the background locus is strong.
Keywords: adaptive divergence; establishment; gene flow; linked selection; local adaptation.
Figures



References
-
- Barton N., 1979. Gene flow past a cline. Heredity 43: 333–339.
-
- Barton N., Bengtsson B. O., 1986. The barrier to genetic exchange between hybridising populations. Heredity 57: 357–376. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

