Recent advances in the study of chloroplast gene expression and its evolution
- PMID: 24611069
- PMCID: PMC3933795
- DOI: 10.3389/fpls.2014.00061
Recent advances in the study of chloroplast gene expression and its evolution
Abstract
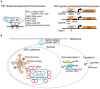
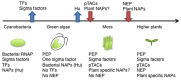
Chloroplasts are semiautonomous organelles which possess their own genome and gene expression system. However, extant chloroplasts contain only limited coding information, and are dependent on a large number of nucleus-encoded proteins. During plant evolution, chloroplasts have lost most of the prokaryotic DNA-binding proteins and transcription regulators that were present in the original endosymbiont. Thus, chloroplasts have a unique hybrid transcription system composed of the remaining prokaryotic components, such as a prokaryotic RNA polymerase as well as nucleus-encoded eukaryotic components. Recent proteomic and transcriptomic analyses have provided insights into chloroplast transcription systems and their evolution. Here, we review chloroplast-specific transcription systems, focusing on the multiple RNA polymerases, eukaryotic transcription regulators in chloroplasts, chloroplast promoters, and the dynamics of chloroplast nucleoids.
Keywords: NEP; PEP; chloroplast; nucleoid; pTAC; transcription.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

