Quorum sensing dependent phenotypes and their molecular mechanisms in Campylobacterales
- PMID: 24611121
- PMCID: PMC3933990
- DOI: 10.1556/EuJMI.2.2012.1.8
Quorum sensing dependent phenotypes and their molecular mechanisms in Campylobacterales
Abstract
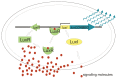
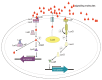
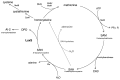
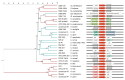
Quorum sensing comprises the mechanism of communication between numerous bacteria via small signalling molecules, termed autoinducers (AI). Using quorum sensing, bacteria can regulate the expression of multiple genes involved in virulence, toxin production, motility, chemotaxis and biofilm formation, thus contributing to adaptation as well as colonisation. The current understanding of the role of quorum sensing in the lifecycle of Campylobacterales is still incomplete. Campylobacterales belong to the class of Epsilonproteobacteria representing a physiologically and ecologically diverse group of bacteria that are rather distinct from the more commonly studied Proteobacteria, such as Escherichia and Salmonella. This review summarises the recent knowledge on distribution and production of AI molecules, as well as possible quorum sensing dependent regulation in the mostly investigated species within the Campylobacterales group: Campylobacter jejuni and Helicobacter pylori.
Keywords: AI-2; Campylobacter; Helicobacter; LuxS; quorum sensing.
Figures

References
-
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
