Bacterial group I introns: mobile RNA catalysts
- PMID: 24612670
- PMCID: PMC3984707
- DOI: 10.1186/1759-8753-5-8
Bacterial group I introns: mobile RNA catalysts
Abstract
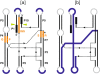
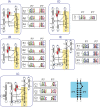
Group I introns are intervening sequences that have invaded tRNA, rRNA and protein coding genes in bacteria and their phages. The ability of group I introns to self-splice from their host transcripts, by acting as ribozymes, potentially renders their insertion into genes phenotypically neutral. Some group I introns are mobile genetic elements due to encoded homing endonuclease genes that function in DNA-based mobility pathways to promote spread to intronless alleles. Group I introns have a limited distribution among bacteria and the current assumption is that they are benign selfish elements, although some introns and homing endonucleases are a source of genetic novelty as they have been co-opted by host genomes to provide regulatory functions. Questions regarding the origin and maintenance of group I introns among the bacteria and phages are also addressed.
Figures



References
-
- Cech TR. Self-splicing of group-I introns. Annu Rev Biochem. 1990;55:599–629. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

