The transcription factor titration effect dictates level of gene expression
- PMID: 24612990
- PMCID: PMC4080642
- DOI: 10.1016/j.cell.2014.02.022
The transcription factor titration effect dictates level of gene expression
Abstract
Models of transcription are often built around a picture of RNA polymerase and transcription factors (TFs) acting on a single copy of a promoter. However, most TFs are shared between multiple genes with varying binding affinities. Beyond that, genes often exist at high copy number-in multiple identical copies on the chromosome or on plasmids or viral vectors with copy numbers in the hundreds. Using a thermodynamic model, we characterize the interplay between TF copy number and the demand for that TF. We demonstrate the parameter-free predictive power of this model as a function of the copy number of the TF and the number and affinities of the available specific binding sites; such predictive control is important for the understanding of transcription and the desire to quantitatively design the output of genetic circuits. Finally, we use these experiments to dynamically measure plasmid copy number through the cell cycle.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


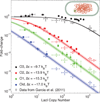

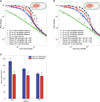

Comment in
-
Target competition: transcription factors enter the limelight.Genome Biol. 2014 Apr 28;15(4):114. doi: 10.1186/gb4174. Genome Biol. 2014. PMID: 25001290 Free PMC article.
References
-
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. - PubMed
-
- Babu MM, Teichmann SA. Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet. 2003;19:75–79. - PubMed
-
- Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, Motifs, and Scaffolds: The Role of Modular Interactions in the Evolution and Wiring of Cell Signaling Circuits. Annu Rev Biochem. 2006;75:655–680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

