Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation
- PMID: 24613091
- PMCID: PMC4210641
- DOI: 10.1016/j.immuni.2014.01.011
Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation
Abstract
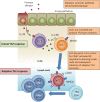
Naive CD4(+) T cell differentiation into distinct subsets of T helper (Th) cells is a pivotal process in the initiation of the adaptive immune response. Allergens predominantly stimulate Th2 cells, causing allergic inflammation. However, why allergens induce Th2 cell differentiation is not well understood. Here we show that group 2 innate lymphoid cells (ILC2s) are required to mount a robust Th2 cell response to the protease-allergen papain. Intranasal administration of papain stimulated ILC2s and Th2 cells, causing allergic lung inflammation and elevated immunoglobulin E titers. This process was severely impaired in ILC2-deficient mice. Whereas interleukin-4 (IL-4) was dispensable for papain-induced Th2 cell differentiation, ILC2-derived IL-13 was critical as it promoted migration of activated lung dendritic cells into the draining lymph node where they primed naive T cells to differentiate into Th2 cells. Papain-induced ILC2 activation and Th2 cell differentiation was IL-33-dependent, suggesting a common pathway in the initiation of Th2 cell responses to allergen.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

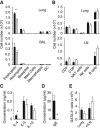
 ) were calculated on days 2 and 21. Data are representative of at least three independent experiments. Mean ± SEM in (B)–(H), mean percent gated in H, ∗ = p ≤ 0.05 ∗∗ = p ≤ 0.01 (two-tailed Student’s t test). See also Figure S1.
) were calculated on days 2 and 21. Data are representative of at least three independent experiments. Mean ± SEM in (B)–(H), mean percent gated in H, ∗ = p ≤ 0.05 ∗∗ = p ≤ 0.01 (two-tailed Student’s t test). See also Figure S1.
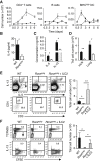

 ). (C) The induction of Th2 cells in WT BMT mice stimulated with papain (▪) was compared to Rorasg/sg BMT mice stimulated with papain (□) or papain + rmIL-13 (
). (C) The induction of Th2 cells in WT BMT mice stimulated with papain (▪) was compared to Rorasg/sg BMT mice stimulated with papain (□) or papain + rmIL-13 ( ). The total numbers of IL-5+IL-13+ CD3+CD4+ Th2 cells in the mLN were quantified on day 6. (D) CFSE-labeled CD4+ OT-II cells were transplanted into WT and Rorasg/sg BMT mice, followed by treatment with papain + OVA. On day 6, mLN cells were analyzed by flow cytometry for CD4+TCRVβ5+CFSE+ OT-II T cell proliferation and intracellular IL-4 and IL-13. (E) WT (▪) and Rorasg/sg (□) were treated as in (D). Some of the latter mice also received rmIL-13 injection (
). The total numbers of IL-5+IL-13+ CD3+CD4+ Th2 cells in the mLN were quantified on day 6. (D) CFSE-labeled CD4+ OT-II cells were transplanted into WT and Rorasg/sg BMT mice, followed by treatment with papain + OVA. On day 6, mLN cells were analyzed by flow cytometry for CD4+TCRVβ5+CFSE+ OT-II T cell proliferation and intracellular IL-4 and IL-13. (E) WT (▪) and Rorasg/sg (□) were treated as in (D). Some of the latter mice also received rmIL-13 injection ( ). OT-II T cells positively stained for intracellular IL-4 or IL-13 were quantified and the total numbers of cytokine positive OT-II in the mLN were calculated. (F) Rorasg/sg BMT mice were transplanted with CFSE-labeled CD4+ OT-II cells plus none, WT, or Il13−/− ILC2s. These mice were treated on days 0 and 1 with papain, and CD3+CD4+TCRVβ5+CFSE+ OT-II T cells in the mLN were analyzed for intracellular IL-4 and IL-13 (plots) on day 6. The total numbers of IL-4+IL-13+ OT-II T cells in the mLN were calculated (bar graph). (G) CFSE-labeled CD4+ OT-II cells were transplanted into WT mice. The mice received intranasal administration of OVA + rmIL-13, OVA alone, or OVA + papain on days 0 and 1. CD3+CD4+TCRVβ5+CFSE+ cells were gated (data not shown) and analyzed for intracellular IL-4 and IL-13 on day 6 (plots). The total numbers of IL-4+IL-13+ OT-II T cells in the mLN were calculated (bar graph). Data are representative of at least three independent experiments. Mean ± SEM in (A)–(C) and (E)–(G), mean percent gated in (D), (F), and (G), ∗ = p ≤ 0.05 ∗∗ = p ≤ 0.01, ns = not significant (two-tailed Student’s t test). See also Figure S5.
). OT-II T cells positively stained for intracellular IL-4 or IL-13 were quantified and the total numbers of cytokine positive OT-II in the mLN were calculated. (F) Rorasg/sg BMT mice were transplanted with CFSE-labeled CD4+ OT-II cells plus none, WT, or Il13−/− ILC2s. These mice were treated on days 0 and 1 with papain, and CD3+CD4+TCRVβ5+CFSE+ OT-II T cells in the mLN were analyzed for intracellular IL-4 and IL-13 (plots) on day 6. The total numbers of IL-4+IL-13+ OT-II T cells in the mLN were calculated (bar graph). (G) CFSE-labeled CD4+ OT-II cells were transplanted into WT mice. The mice received intranasal administration of OVA + rmIL-13, OVA alone, or OVA + papain on days 0 and 1. CD3+CD4+TCRVβ5+CFSE+ cells were gated (data not shown) and analyzed for intracellular IL-4 and IL-13 on day 6 (plots). The total numbers of IL-4+IL-13+ OT-II T cells in the mLN were calculated (bar graph). Data are representative of at least three independent experiments. Mean ± SEM in (A)–(C) and (E)–(G), mean percent gated in (D), (F), and (G), ∗ = p ≤ 0.05 ∗∗ = p ≤ 0.01, ns = not significant (two-tailed Student’s t test). See also Figure S5.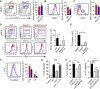
 ), or control (□) were analyzed for absolute numbers of activated DCs in mLNs on day 3 as in (D). (F) Traceable DQ-OVA was injected together with papain into WT (red) or Rorasg/sg (blue) BMT mice, followed by an analysis for DQ-positive CD11c+MHCIIhi DCs in the lung and mLN on day 3 (histograms), and the total numbers of DQ+ DCs were calculated (bar graph). (G) Lung tissue explants were made from papain-stimulated WT, Il13−/−, or Il13−/− + rmIL-13 mice on day 3. Tissue explants were cultured in transwell inserts and exposed to a CCL21 chemokine gradient for 14 hr. WT lung explants without a CCL21 gradient were used as control. The total number of activated DCs was counted in both top and bottom compartments by flow cytometry, followed by a calculation of percent DC migration. (H) Lung tissue explants from naive WT mice were stimulated ex vivo with papain for 14 hr in the presence of anti-IL-13 neutralizing antibody ± EP4-agonist or PBS control. The percent of DC migration toward a CCL21 chemokine gradient was calculated. (I) Lung explants were prepared, treated with papain in the presence of PBS (control), anti-IL-13 or EP4 antagonist, and DC migration was analyzed as in (H). Data are representative of at least three independent experiments. Mean ± SEM in (A)–(I), mean percent gated in (A) and (D), ns = not significant ∗ = p ≤ 0.05, ∗∗ = p ≤ 0.01 ∗∗∗ = p ≤ 0.001 (two-tailed Student’s t test). See also Figure S6.
), or control (□) were analyzed for absolute numbers of activated DCs in mLNs on day 3 as in (D). (F) Traceable DQ-OVA was injected together with papain into WT (red) or Rorasg/sg (blue) BMT mice, followed by an analysis for DQ-positive CD11c+MHCIIhi DCs in the lung and mLN on day 3 (histograms), and the total numbers of DQ+ DCs were calculated (bar graph). (G) Lung tissue explants were made from papain-stimulated WT, Il13−/−, or Il13−/− + rmIL-13 mice on day 3. Tissue explants were cultured in transwell inserts and exposed to a CCL21 chemokine gradient for 14 hr. WT lung explants without a CCL21 gradient were used as control. The total number of activated DCs was counted in both top and bottom compartments by flow cytometry, followed by a calculation of percent DC migration. (H) Lung tissue explants from naive WT mice were stimulated ex vivo with papain for 14 hr in the presence of anti-IL-13 neutralizing antibody ± EP4-agonist or PBS control. The percent of DC migration toward a CCL21 chemokine gradient was calculated. (I) Lung explants were prepared, treated with papain in the presence of PBS (control), anti-IL-13 or EP4 antagonist, and DC migration was analyzed as in (H). Data are representative of at least three independent experiments. Mean ± SEM in (A)–(I), mean percent gated in (A) and (D), ns = not significant ∗ = p ≤ 0.05, ∗∗ = p ≤ 0.01 ∗∗∗ = p ≤ 0.001 (two-tailed Student’s t test). See also Figure S6.Comment in
-
Innate immune cells to the help.Immunity. 2014 Mar 20;40(3):313-4. doi: 10.1016/j.immuni.2014.02.010. Immunity. 2014. PMID: 24656044
-
Innate lymphoid cells: breathing into allergic inflammation.Nat Rev Immunol. 2014 May;14(5):281. doi: 10.1038/nri3668. Epub 2014 Apr 11. Nat Rev Immunol. 2014. PMID: 24722478 No abstract available.
References
-
- Barlow J.L., Bellosi A., Hardman C.S., Drynan L.F., Wong S.H. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J. Allergy Clin. Immunol. 2012;129 191–8.e1–4. - PubMed
-
- Besnard A.-G., Togbe D., Guillou N., Erard F., Quesniaux V., Ryffel B. IL-33-activated dendritic cells are critical for allergic airway inflammation. Eur. J. Immunol. 2011;41:1675–1686. - PubMed
-
- Bromley S.K., Mempel T.R., Luster A.D. Orchestrating the orchestrators: chemokines in control of T cell traffic. Nat. Immunol. 2008;9:970–980. - PubMed
-
- Finkelman F.D., Katona I.M., Urban J.F., Jr., Holmes J., Ohara J., Tung A.S., Sample J.V., Paul W.E. IL-4 is required to generate and sustain in vivo IgE responses. J. Immunol. 1988;141:2335–2341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

