Identification of transcriptional and metabolic programs related to mammalian cell size
- PMID: 24613310
- PMCID: PMC3991852
- DOI: 10.1016/j.cub.2014.01.071
Identification of transcriptional and metabolic programs related to mammalian cell size
Abstract
Background: Regulation of cell size requires coordination of growth and proliferation. Conditional loss of cyclin-dependent kinase 1 in mice permits hepatocyte growth without cell division, allowing us to study cell size in vivo using transcriptomics and metabolomics.
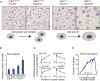
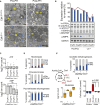
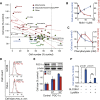
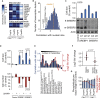
Results: Larger cells displayed increased expression of cytoskeletal genes but unexpectedly repressed expression of many genes involved in mitochondrial functions. This effect appears to be cell autonomous because cultured Drosophila cells induced to increase cell size displayed a similar gene-expression pattern. Larger hepatocytes also displayed a reduction in the expression of lipogenic transcription factors, especially sterol-regulatory element binding proteins. Inhibition of mitochondrial functions and lipid biosynthesis, which is dependent on mitochondrial metabolism, increased the cell size with reciprocal effects on cell proliferation in several cell lines.
Conclusions: We uncover that large cell-size increase is accompanied by downregulation of mitochondrial gene expression, similar to that observed in diabetic individuals. Mitochondrial metabolism and lipid synthesis are used to couple cell size and cell proliferation. This regulatory mechanism may provide a possible mechanism for sensing metazoan cell size.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Comment in
-
Cell growth: when less means more.Curr Biol. 2014 Mar 31;24(7):R283-5. doi: 10.1016/j.cub.2014.02.044. Curr Biol. 2014. PMID: 24698379
References
-
- Lloyd A.C. The regulation of cell size. Cell. 2013;154:1194–1205. - PubMed
-
- Jorgensen P., Tyers M. How cells coordinate growth and division. Curr. Biol. 2004;14:R1014–R1027. - PubMed
-
- Marguerat S., Bähler J. Coordinating genome expression with cell size. Trends Genet. 2012;28:560–565. - PubMed
-
- Jorgensen P., Nishikawa J.L., Breitkreutz B.J., Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

