Non-homologous end joining often uses microhomology: implications for alternative end joining
- PMID: 24613510
- PMCID: PMC4440676
- DOI: 10.1016/j.dnarep.2014.02.006
Non-homologous end joining often uses microhomology: implications for alternative end joining
Abstract
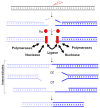
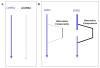
Artemis and PALF (also called APLF) appear to be among the primary nucleases involved in non-homologous end joining (NHEJ) and responsible for most nucleolytic end processing in NHEJ. About 60% of NHEJ events show an alignment of the DNA ends that use 1 or 2bp of microhomology (MH) between the two DNA termini. Thus, MH is a common feature of NHEJ. For most naturally occurring human chromosomal deletions (e.g., after oxidative damage or radiation) and translocations, such as those seen in human neoplasms and as well as inherited chromosomal structural variations, MH usage occurs at a frequency that is typical of NHEJ, and does not suggest major involvement of alternative pathways that require more extensive MH. Though we mainly focus on human NHEJ at double-strand breaks, comparison on these points to other eukaryotes, primarily S. cerevisiae, is informative.
Keywords: Double-strand break repair, Lymphoma, Chromosomal rearrangements, V(D)J recombination, Class switch recombination.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures


References
-
- Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. - PubMed
-
- Moshous D, Li L, Chasseval R, Philippe N, Jabado N, Cowan MJ, Fischer A, Villartay J-Pd. A new gene involved in DNA double-strand break repair and V(D)J recombination is located on human chromosome 10p. Hum Mol Gen. 2000;9:583–588. - PubMed
-
- Li L, Moshous D, Zhou Y, Wang J, Xie G, Salido E, Hu D, Villartay JPd, Cowan MJ. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J Immunol. 2002;168:6323–6329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

