Folding of pig gastric mucin non-glycosylated domains: a discrete molecular dynamics study
- PMID: 24615227
- PMCID: PMC3473135
- DOI: 10.1007/s10867-012-9280-x
Folding of pig gastric mucin non-glycosylated domains: a discrete molecular dynamics study
Abstract
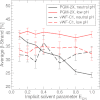
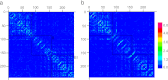
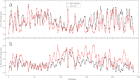
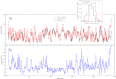
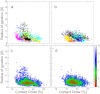
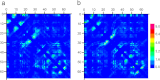
Mucin glycoproteins consist of tandem-repeating glycosylated regions flanked by non-repetitive protein domains with little glycosylation. These non-repetitive domains are involved in polymerization of mucin and play an important role in the pH-dependent gelation of gastric mucin, which is essential for protecting the stomach from autodigestion. We examine folding of the non-repetitive sequence of PGM-2X (242 amino acids) and the von Willebrand factor vWF-C1 domain (67 amino acids) at neutral and low pH using discrete molecular dynamics (DMD) in an implicit solvent combined with a four-bead peptide model. Using the same implicit solvent parameters, folding of both domains is simulated at neutral and low pH. In contrast to vWF-C1, PGM-2X folding is strongly affected by pH as indicated by changes in the contact order, radius of gyration, free-energy landscape, and the secondary structure. Whereas the free-energy landscape of vWF-C1 shows a single minimum at both neutral and low pH, the free-energy landscape of PGM-2X is characterized by multiple minima that are more numerous and shallower at low pH. Detailed structural analysis shows that PGM-2X partially unfolds at low pH. This partial unfolding is facilitated by the C-terminal region GLU236-PRO242, which loses contact with the rest of the domain due to effective "mean-field" repulsion among highly positively charged N- and C-terminal regions. Consequently, at low pH, hydrophobic amino acids are more exposed to the solvent. In vWF-C1, low pH induces some structural changes, including an increased exposure of CYS at position 67, but these changes are small compared to those found in PGM-2X. For PGM-2X, the DMD-derived average β-strand propensity increases from 0.26 ± 0.01 at neutral pH to 0.38 ± 0.01 at low pH. For vWF-C1, the DMD-derived average β-strand propensity is 0.32 ± 0.02 at neutral pH and 0.35 ± 0.02 at low pH. The DMD-derived structural information provides insight into pH-induced changes in the folding of two distinct mucin domains and suggests plausible mechanisms of the aggregation/gelation of mucin.
Figures


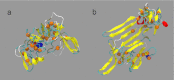
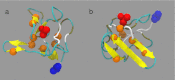
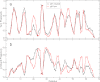
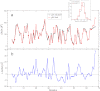
References
-
- Bansil R, Turner BS. Mucin structure, aggregation, physiological functions and biomedical applications. Curr. Opin. Colloid Interface Sci. 2006;11:164–170. doi: 10.1016/j.cocis.2005.11.001. - DOI
-
- Turner, B.S.: Expression of cysteine-rich pig gastric mucin (MUC5AC) domains in Pichia pastoris and their pH-dependent properties. Dissertation, Boston University (2012)
-
- Bhaskar KR, Gong D, Bansil R, Pajevic S, Hamilton JA, Turner BS, Lamont JT. Profound increase in viscosity and aggregation of pig gastric mucin at low pH. Am. J. Physiol. 1991;261:827–833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

