After embedding in membranes antiapoptotic Bcl-XL protein binds both Bcl-2 homology region 3 and helix 1 of proapoptotic Bax protein to inhibit apoptotic mitochondrial permeabilization
- PMID: 24616095
- PMCID: PMC4002096
- DOI: 10.1074/jbc.M114.552562
After embedding in membranes antiapoptotic Bcl-XL protein binds both Bcl-2 homology region 3 and helix 1 of proapoptotic Bax protein to inhibit apoptotic mitochondrial permeabilization
Abstract
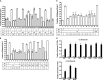
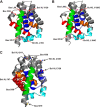
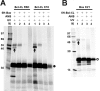
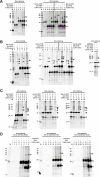
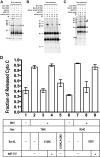
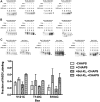
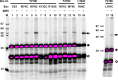
Bcl-XL binds to Bax, inhibiting Bax oligomerization required for mitochondrial outer membrane permeabilization (MOMP) during apoptosis. How Bcl-XL binds to Bax in the membrane is not known. Here, we investigated the structural organization of Bcl-XL·Bax complexes formed in the MOM, including the binding interface and membrane topology, using site-specific cross-linking, compartment-specific labeling, and computational modeling. We found that one heterodimer interface is formed by a specific interaction between the Bcl-2 homology 1-3 (BH1-3) groove of Bcl-XL and the BH3 helix of Bax, as defined previously by the crystal structure of a truncated Bcl-XL protein and a Bax BH3 peptide (Protein Data Bank entry 3PL7). We also discovered a novel interface in the heterodimer formed by equivalent interactions between the helix 1 regions of Bcl-XL and Bax when their helical axes are oriented either in parallel or antiparallel. The two interfaces are located on the cytosolic side of the MOM, whereas helix 9 of Bcl-XL is embedded in the membrane together with helices 5, 6, and 9 of Bax. Formation of the helix 1·helix 1 interface partially depends on the formation of the groove·BH3 interface because point mutations in the latter interface and the addition of ABT-737, a groove-binding BH3 mimetic, blocked the formation of both interfaces. The mutations and ABT-737 also prevented Bcl-XL from inhibiting Bax oligomerization and subsequent MOMP, suggesting that the structural organization in which interactions at both interfaces contribute to the overall stability and functionality of the complex represents antiapoptotic Bcl-XL·Bax complexes in the MOM.
Keywords: Bax; Bcl-2 Proteins; Bcl-XL; Membrane Proteins; Mitochondrial Apoptosis; Protein Cross-linking.
Figures



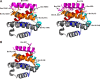
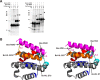



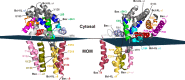
References
-
- Kvansakul M., Yang H., Fairlie W. D., Czabotar P. E., Fischer S. F., Perugini M. A., Huang D. C., Colman P. M. (2008) Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ. 15, 1564–1571 - PubMed
-
- Youle R. J., Strasser A. (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

