Specific inhibition of diverse pathogens in human cells by synthetic microRNA-like oligonucleotides inferred from RNAi screens
- PMID: 24616511
- PMCID: PMC3970520
- DOI: 10.1073/pnas.1402353111
Specific inhibition of diverse pathogens in human cells by synthetic microRNA-like oligonucleotides inferred from RNAi screens
Abstract
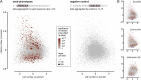
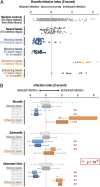
Systematic genetic perturbation screening in human cells remains technically challenging. Typically, large libraries of chemically synthesized siRNA oligonucleotides are used, each designed to degrade a specific cellular mRNA via the RNA interference (RNAi) mechanism. Here, we report on data from three genome-wide siRNA screens, conducted to uncover host factors required for infection of human cells by two bacterial and one viral pathogen. We find that the majority of phenotypic effects of siRNAs are unrelated to the intended "on-target" mechanism, defined by full complementarity of the 21-nt siRNA sequence to a target mRNA. Instead, phenotypes are largely dictated by "off-target" effects resulting from partial complementarity of siRNAs to multiple mRNAs via the "seed" region (i.e., nucleotides 2-8), reminiscent of the way specificity is determined for endogenous microRNAs. Quantitative analysis enabled the prediction of seeds that strongly and specifically block infection, independent of the intended on-target effect. This prediction was confirmed experimentally by designing oligos that do not have any on-target sequence match at all, yet can strongly reproduce the predicted phenotypes. Our results suggest that published RNAi screens have primarily, and unintentionally, screened the sequence space of microRNA seeds instead of the intended on-target space of protein-coding genes. This helps to explain why previously published RNAi screens have exhibited relatively little overlap. Our analysis suggests a possible way of identifying "seed reagents" for controlling phenotypes of interest and establishes a general strategy for extracting valuable untapped information from past and future RNAi screens.
Keywords: antimicrobials; high-throughput RNAi screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


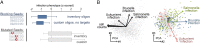
Comment in
-
Techniques and applications: RNAi 'off-targets' pathogen infection.Nat Rev Microbiol. 2014 May;12(5):314. doi: 10.1038/nrmicro3257. Epub 2014 Mar 24. Nat Rev Microbiol. 2014. PMID: 24686412 No abstract available.
-
Off-targets in RNAi screens.Nat Methods. 2014 May;11(5):480. doi: 10.1038/nmeth.2958. Nat Methods. 2014. PMID: 24820375 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

