Revealing insect herbivory-induced phenolamide metabolism: from single genes to metabolic network plasticity analysis
- PMID: 24617849
- PMCID: PMC5140026
- DOI: 10.1111/tpj.12503
Revealing insect herbivory-induced phenolamide metabolism: from single genes to metabolic network plasticity analysis
Abstract
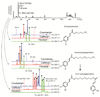
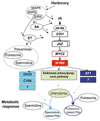
The phenylpropanoid metabolic space comprises a network of interconnected metabolic branches that contribute to the biosynthesis of a large array of compounds with functions in plant development and stress adaptation. During biotic challenges, such as insect attack, a major rewiring of gene networks associated with phenylpropanoid metabolism is observed. This rapid reconfiguration of gene expression allows prioritized production of metabolites that help the plant solve ecological problems. Phenolamides are a group of phenolic derivatives that originate from diversion of hydroxycinnamoyl acids from the main phenylpropanoid pathway after N-acyltransferase-dependent conjugation to polyamines or aryl monoamines. These structurally diverse metabolites are abundant in the reproductive organs of many plants, and have recently been shown to play roles as induced defenses in vegetative tissues. In the wild tobacco, Nicotiana attenuata, in which herbivory-induced regulation of these metabolites has been studied, rapid elevations of the levels of phenolamides that function as induced defenses result from a multi-hormonal signaling network that re-shapes connected metabolic pathways. In this review, we summarize recent findings in the regulation of phenolamides obtained by mass spectrometry-based metabolomics profiling, and outline a conceptual framework for gene discovery in this pathway. We also introduce a multifactorial approach that is useful in deciphering metabolic pathway reorganizations among tissues in response to stress.
Keywords: N-acyltransferase; Nicotiana attenuata; metabolomics; phenolamides; phenylpropanoid pathway; self-organizing maps; systems biology.
© 2014 The Authors The Plant Journal © 2014 John Wiley & Sons Ltd.
Figures




References
-
- Agrawal AA, Hastings AP, Johnson MT, Maron JL, Salminen J-P. Insect herbivores drive real-time ecological and evolutionary change in plant populations. Science. 338:113–116. - PubMed
-
- Bassard JE, Ullmann P, Bernier F, Werck-Reichhart D. Phenolamides: Bridging polyamines to the phenolic metabolism. Phytochemistry. 2010;71:1808–1824. - PubMed
-
- Bittner M, Meltzer P, Trent J. Data analysis and integration: of steps and arrows. Nat Genet. 1999;22:213–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

