Paleo-evolutionary plasticity of plant disease resistance genes
- PMID: 24617999
- PMCID: PMC4234491
- DOI: 10.1186/1471-2164-15-187
Paleo-evolutionary plasticity of plant disease resistance genes
Abstract
Background: The recent access to a large set of genome sequences, combined with a robust evolutionary scenario of modern monocot (i.e. grasses) and eudicot (i.e. rosids) species from their founder ancestors, offered the opportunity to gain insights into disease resistance genes (R-genes) evolutionary plasticity.
Results: We unravel in the current article (i) a R-genes repertoire consisting in 7883 for monocots and 15758 for eudicots, (ii) a contrasted R-genes conservation with 23.8% for monocots and 6.6% for dicots, (iii) a minimal ancestral founder pool of 384 R-genes for the monocots and 150 R-genes for the eudicots, (iv) a general pattern of organization in clusters accounting for more than 60% of mapped R-genes, (v) a biased deletion of ancestral duplicated R-genes between paralogous blocks possibly compensated by clusterization, (vi) a bias in R-genes clusterization where Leucine-Rich Repeats act as a 'glue' for domain association, (vii) a R-genes/miRNAs interome enriched toward duplicated R-genes.
Conclusions: Together, our data may suggest that R-genes family plasticity operated during plant evolution (i) at the structural level through massive duplicates loss counterbalanced by massive clusterization following polyploidization; as well as at (ii) the regulation level through microRNA/R-gene interactions acting as a possible source of functional diploidization of structurally retained R-genes duplicates. Such evolutionary shuffling events leaded to CNVs (i.e. Copy Number Variation) and PAVs (i.e. Presence Absence Variation) between related species operating in the decay of R-genes colinearity between plant species.
Figures
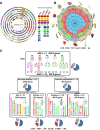
 illustrates non-significant differences;
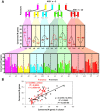
illustrates non-significant differences;  illustrates significant differences based on permutation test with P-value < 0.05, see Methods section). Distribution pattern of conserved R-genes in modern chromosome pairs is shown in rice, Brachypodium, sorghum and maize. (B) Illustration of the correlation between the number of observed R-genes in cluster (x-axis) and the total number of conserved R-genes (y-axis) characterized in the sensitive (red dot and curve) and dominant (black dot and curve) chromosomal blocks.
illustrates significant differences based on permutation test with P-value < 0.05, see Methods section). Distribution pattern of conserved R-genes in modern chromosome pairs is shown in rice, Brachypodium, sorghum and maize. (B) Illustration of the correlation between the number of observed R-genes in cluster (x-axis) and the total number of conserved R-genes (y-axis) characterized in the sensitive (red dot and curve) and dominant (black dot and curve) chromosomal blocks.

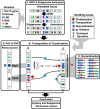
References
-
- Pallas V, Garcia JA. How do plant viruses induce disease? Interactions and interference with host components. J Gen Virol. 2011;92(Pt 12):2691–2705. - PubMed
-
- Soriano IR, Riley IT, Potter MJ, Bowers WS. Phytoecdysteroids: a novel defense against plant-parasitic nematodes. J Chem Ecol. 2004;30(10):1885–1899. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

