p53 activity is selectively licensed in the Drosophila stem cell compartment
- PMID: 24618896
- PMCID: PMC3949305
- DOI: 10.7554/eLife.01530
p53 activity is selectively licensed in the Drosophila stem cell compartment
Abstract
Oncogenic stress provokes tumor suppression by p53 but the extent to which this regulatory axis is conserved remains unknown. Using a biosensor to visualize p53 action, we find that Drosophila p53 is selectively active in gonadal stem cells after exposure to stressors that destabilize the genome. Similar p53 activity occurred in hyperplastic growths that were triggered either by the Ras(V12) oncoprotein or by failed differentiation programs. In a model of transient sterility, p53 was required for the recovery of fertility after stress, and entry into the cell cycle was delayed in p53(-) stem cells. Together, these observations establish that the stem cell compartment of the Drosophila germline is selectively licensed for stress-induced activation of the p53 regulatory network. Furthermore, the findings uncover ancestral links between p53 and aberrant proliferation that are independent of DNA breaks and predate evolution of the ARF/Mdm2 axis. DOI: http://dx.doi.org/10.7554/eLife.01530.001.
Keywords: Drosophila; biosensor; germline; oncogenic stress; p53; stem cells.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
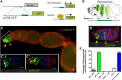
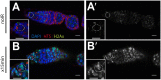
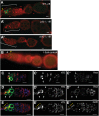
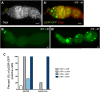


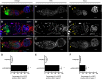
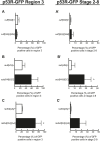

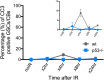

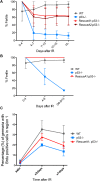



References
-
- Chen D, McKearin DM. 2003. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130:1159–1170 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

