Distinguishing Between Reservoir Exposure and Human-to-Human Transmission for Emerging Pathogens Using Case Onset Data
- PMID: 24619563
- PMCID: PMC3946006
- DOI: 10.1371/currents.outbreaks.e1473d9bfc99d080ca242139a06c455f
Distinguishing Between Reservoir Exposure and Human-to-Human Transmission for Emerging Pathogens Using Case Onset Data
Abstract
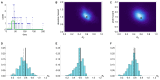
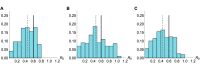
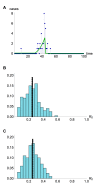
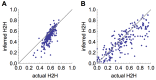
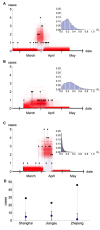
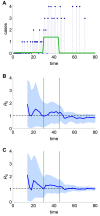
Pathogens such as MERS-CoV, influenza A/H5N1 and influenza A/H7N9 are currently generating sporadic clusters of spillover human cases from animal reservoirs. The lack of a clear human epidemic suggests that the basic reproductive number R0 is below or very close to one for all three infections. However, robust cluster-based estimates for low R0 values are still desirable so as to help prioritise scarce resources between different emerging infections and to detect significant changes between clusters and over time. We developed an inferential transmission model capable of distinguishing the signal of human-to-human transmission from the background noise of direct spillover transmission (e.g. from markets or farms). By simulation, we showed that our approach could obtain unbiased estimates of R0, even when the temporal trend in spillover exposure was not fully known, so long as the serial interval of the infection and the timing of a sudden drop in spillover exposure were known (e.g. day of market closure). Applying our method to data from the three largest outbreaks of influenza A/H7N9 outbreak in China in 2013, we found evidence that human-to-human transmission accounted for 13% (95% credible interval 1%-32%) of cases overall. We estimated R0 for the three clusters to be: 0.19 in Shanghai (0.01-0.49), 0.29 in Jiangsu (0.03-0.73); and 0.03 in Zhejiang (0.00-0.22). If a reliable temporal trend for the spillover hazard could be estimated, for example by implementing widespread routine sampling in sentinel markets, it should be possible to estimate sub-critical values of R0 even more accurately. Should a similar strain emerge with R0>1, these methods could give a real-time indication that sustained transmission is occurring with well-characterised uncertainty.
Keywords: H7N9; Influenza; infectious diseases; statistical inference; zoonoses.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

