PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics
- PMID: 24619595
- PMCID: PMC4056586
- DOI: 10.1002/ajmg.c.31391
PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics
Abstract
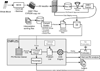
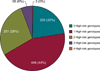
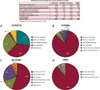
Pharmacogenetics is frequently cited as an area for initial focus of the clinical implementation of genomics. Through the PG4KDS protocol, St. Jude Children's Research Hospital pre-emptively genotypes patients for 230 genes using the Affymetrix Drug Metabolizing Enzymes and Transporters (DMET) Plus array supplemented with a CYP2D6 copy number assay. The PG4KDS protocol provides a rational, stepwise process for implementing gene/drug pairs, organizing data, and obtaining consent from patients and families. Through August 2013, 1,559 patients have been enrolled, and four gene tests have been released into the electronic health record (EHR) for clinical implementation: TPMT, CYP2D6, SLCO1B1, and CYP2C19. These genes are coupled to 12 high-risk drugs. Of the 1,016 patients with genotype test results available, 78% of them had at least one high-risk (i.e., actionable) genotype result placed in their EHR. Each diplotype result released to the EHR is coupled with an interpretive consult that is created in a concise, standardized format. To support-gene based prescribing at the point of care, 55 interruptive clinical decision support (CDS) alerts were developed. Patients are informed of their genotyping result and its relevance to their medication use through a letter. Key elements necessary for our successful implementation have included strong institutional support, a knowledgeable clinical laboratory, a process to manage any incidental findings, a strategy to educate clinicians and patients, a process to return results, and extensive use of informatics, especially CDS. Our approach to pre-emptive clinical pharmacogenetics has proven feasible, clinically useful, and scalable.
Keywords: clinical decision support; electronic health record; personalized medicine; pharmacogenetics; pharmacogenomics.
© 2014 Wiley Periodicals, Inc.
Conflict of interest statement
Figures
References
-
- Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, Baker DK, Kornegay NM, Yang W, Cross SJ, Howard SC, Freimuth RR, Evans WE, Broeckel U, Relling MV, Hoffman JM. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2013 [Epub ahead of print]. - PMC - PubMed
-
- Cheok MH, Evans WE. Acute lymphoblastic leukaemia: a model for the pharmacogenomics of cancer therapy. Nat Rev Cancer. 2006;6:117–129. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

