Development of MHC-Linked Microsatellite Markers in the Domestic Cat and Their Use to Evaluate MHC Diversity in Domestic Cats, Cheetahs, and Gir Lions
- PMID: 24620003
- PMCID: PMC4048552
- DOI: 10.1093/jhered/esu017
Development of MHC-Linked Microsatellite Markers in the Domestic Cat and Their Use to Evaluate MHC Diversity in Domestic Cats, Cheetahs, and Gir Lions
Abstract
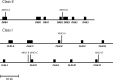
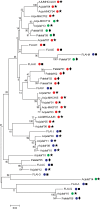
Diversity within the major histocompatibility complex (MHC) reflects the immunological fitness of a population. MHC-linked microsatellite markers provide a simple and an inexpensive method for studying MHC diversity in large-scale studies. We have developed 6 MHC-linked microsatellite markers in the domestic cat and used these, in conjunction with 5 neutral microsatellites, to assess MHC diversity in domestic mixed breed (n = 129) and purebred Burmese (n = 61) cat populations in Australia. The MHC of outbred Australian cats is polymorphic (average allelic richness = 8.52), whereas the Burmese population has significantly lower MHC diversity (average allelic richness = 6.81; P < 0.01). The MHC-linked microsatellites along with MHC cloning and sequencing demonstrated moderate MHC diversity in cheetahs (n = 13) and extremely low diversity in Gir lions (n = 13). Our MHC-linked microsatellite markers have potential future use in diversity and disease studies in other populations and breeds of cats as well as in wild felid species.
Keywords: major histocompatibility complex; Acinonyx jubatus; Felis catus; Panthera leo.
© The American Genetic Association 2014. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Abbott I. 2002. Origin and spread of the cat, Felis catus, on mainland Australia, with a discussion of the magnitude of its early impact on native fauna. Wildlife Res. 29:51–74.
-
- Abbott I. 2008. The spread of the cat, Felis catus, in Australia: re-examination of the current conceptual model with additional information. Conservation Science W. Aust. 7:1–17.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

