Natural antisense transcripts and long non-coding RNA in Neurospora crassa
- PMID: 24621812
- PMCID: PMC3951366
- DOI: 10.1371/journal.pone.0091353
Natural antisense transcripts and long non-coding RNA in Neurospora crassa
Abstract
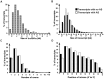
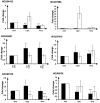
The prevalence of long non-coding RNAs (lncRNA) and natural antisense transcripts (NATs) has been reported in a variety of organisms. While a consensus has yet to be reached on their global importance, an increasing number of examples have been shown to be functional, regulating gene expression at the transcriptional and post-transcriptional level. Here, we use RNA sequencing data from the ABI SOLiD platform to identify lncRNA and NATs obtained from samples of the filamentous fungus Neurospora crassa grown under different light and temperature conditions. We identify 939 novel lncRNAs, of which 477 are antisense to annotated genes. Across the whole dataset, the extent of overlap between sense and antisense transcripts is large: 371 sense/antisense transcripts are complementary over 500 nts or more and 236 overlap by more than 1000 nts. Most prevalent are 3' end overlaps between convergently transcribed sense/antisense pairs, but examples of divergently transcribed pairs and nested transcripts are also present. We confirm the expression of a subset of sense/antisense transcript pairs by qPCR. We examine the size, types of overlap and expression levels under the different environmental stimuli of light and temperature, and identify 11 lncRNAs that are up-regulated in response to light. We also find differences in transcript length and the position of introns between protein-coding transcripts that have antisense expression and transcripts with no antisense expression. These results demonstrate the ability of N. crassa lncRNAs and NATs to be regulated by different environmental stimuli and provide the scope for further investigation into the function of NATs.
Conflict of interest statement
Figures


Similar articles
-
The coding and noncoding transcriptome of Neurospora crassa.BMC Genomics. 2017 Dec 19;18(1):978. doi: 10.1186/s12864-017-4360-8. BMC Genomics. 2017. PMID: 29258423 Free PMC article.
-
Role for antisense RNA in regulating circadian clock function in Neurospora crassa.Nature. 2003 Feb 27;421(6926):948-52. doi: 10.1038/nature01427. Nature. 2003. PMID: 12607002
-
Natural antisense transcripts as versatile regulators of gene expression.Nat Rev Genet. 2024 Oct;25(10):730-744. doi: 10.1038/s41576-024-00723-z. Epub 2024 Apr 17. Nat Rev Genet. 2024. PMID: 38632496 Review.
-
Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis.Genome Res. 2014 Mar;24(3):444-53. doi: 10.1101/gr.165555.113. Epub 2014 Jan 8. Genome Res. 2014. PMID: 24402519 Free PMC article.
-
[Non-coding Natural Antisense RNA: Mechanisms of Action in the Regulation of Target Gene Expression and Its Clinical Implications].Yakugaku Zasshi. 2020;140(5):687-700. doi: 10.1248/yakushi.20-00002. Yakugaku Zasshi. 2020. PMID: 32378673 Review. Japanese.
Cited by
-
Genome Wide Identification and Functional Prediction of Long Non-Coding RNAs Responsive to Sclerotinia sclerotiorum Infection in Brassica napus.PLoS One. 2016 Jul 7;11(7):e0158784. doi: 10.1371/journal.pone.0158784. eCollection 2016. PLoS One. 2016. PMID: 27388760 Free PMC article.
-
The Unexpected Tuners: Are LncRNAs Regulating Host Translation during Infections?Toxins (Basel). 2017 Nov 3;9(11):357. doi: 10.3390/toxins9110357. Toxins (Basel). 2017. PMID: 29469820 Free PMC article.
-
The Effect of Blue Light on the Production of Citrinin in Monascus purpureus M9 by Regulating the mraox Gene through lncRNA AOANCR.Toxins (Basel). 2019 Sep 13;11(9):536. doi: 10.3390/toxins11090536. Toxins (Basel). 2019. PMID: 31540336 Free PMC article.
-
ARHGAP42 promotes cell migration and invasion involving PI3K/Akt signaling pathway in nasopharyngeal carcinoma.Cancer Med. 2018 Aug;7(8):3862-3874. doi: 10.1002/cam4.1552. Epub 2018 Jun 24. Cancer Med. 2018. PMID: 29936709 Free PMC article.
-
Non-coding RNAs as Emerging Regulators of Neural Injury Responses and Regeneration.Neurosci Bull. 2016 Jun;32(3):253-64. doi: 10.1007/s12264-016-0028-7. Epub 2016 Apr 1. Neurosci Bull. 2016. PMID: 27037691 Free PMC article. Review.
References
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

