Transcriptome profiling of a multiple recurrent muscle-invasive urothelial carcinoma of the bladder by deep sequencing
- PMID: 24622401
- PMCID: PMC3951401
- DOI: 10.1371/journal.pone.0091466
Transcriptome profiling of a multiple recurrent muscle-invasive urothelial carcinoma of the bladder by deep sequencing
Abstract
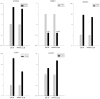
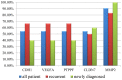
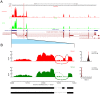
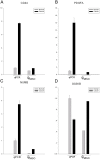
Urothelial carcinoma of the bladder (UCB) is one of the commonly diagnosed cancers in the world. The UCB has the highest rate of recurrence of any malignancy. A genome-wide screening of transcriptome dysregulation between cancer and normal tissue would provide insight into the molecular basis of UCB recurrence and is a key step to discovering biomarkers for diagnosis and therapeutic targets. Compared with microarray technology, which is commonly used to identify expression level changes, the recently developed RNA-seq technique has the ability to detect other abnormal regulations in the cancer transcriptome, such as alternative splicing. In this study, we performed high-throughput transcriptome sequencing at ∼50× coverage on a recurrent muscle-invasive cisplatin-resistance UCB tissue and the adjacent non-tumor tissue. The results revealed cancer-specific differentially expressed genes between the tumor and non-tumor tissue enriched in the cell adhesion molecules, focal adhesion and ECM-receptor interaction pathway. Five dysregulated genes, including CDH1, VEGFA, PTPRF, CLDN7, and MMP2 were confirmed by Real time qPCR in the sequencing samples and the additional eleven samples. Our data revealed that more than three hundred genes showed differential splicing patterns between tumor tissue and non-tumor tissue. Among these genes, we filtered 24 cancer-associated alternative splicing genes with differential exon usage. The findings from RNA-Seq were validated by Real time qPCR for CD44, PDGFA, NUMB, and LPHN2. This study provides a comprehensive survey of the UCB transcriptome, which provides better insight into the complexity of regulatory changes during recurrence and metastasis.
Conflict of interest statement
Figures
References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. (2008) Cancer statistics, 2008. CA Cancer J Clin 58: 71–96. - PubMed
-
- Botteman MF, Pashos CL, Redaelli A, Laskin B, Hauser R (2003) The health economics of bladder cancer: a comprehensive review of the published literature. Pharmacoeconomics 21: 1315–1330. - PubMed
-
- Hayne D, Arya M, Quinn MJ, Babb PJ, Beacock CJ, et al. (2004) Current trends in bladder cancer in England and Wales. J Urol 172: 1051–1055. - PubMed
-
- Williams SG, Stein JP (2004) Molecular pathways in bladder cancer. Urological Research 32: 373–385. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

