Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells
- PMID: 24623245
- PMCID: PMC4407362
- DOI: 10.1007/978-1-62703-739-6_31
Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells
Abstract
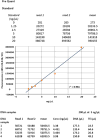
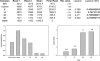
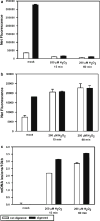
In this chapter, we describe a gene-specific quantitative PCR (QPCR)-based assay for the measurement of DNA damage, using amplification of long DNA targets. This assay has been used extensively to measure the integrity of both nuclear and mitochondrial genomes exposed to different genotoxins and has proven to be particularly valuable in identifying reactive oxygen species-mediated mitochondrial DNA damage. QPCR can be used to quantify both the formation of DNA damage as well as the kinetics of damage removal. One of the main strengths of the assay is that it permits monitoring the integrity of mtDNA directly from total cellular DNA without the need for isolating mitochondria or a separate step of mitochondrial DNA purification. Here we discuss advantages and limitations of using QPCR to assay DNA damage in mammalian cells. In addition, we give a detailed protocol of the QPCR assay that helps facilitate its successful deployment in any molecular biology laboratory.
Figures
References
-
- Van Houten B, Cheng S, Chen Y. Measuring DNA damage and repair in human genes using quantitative amplification of long targets from nanogram quantities of DNA. Mutat Res. 2000;460:81–94. - PubMed
-
- Van Houten B, Chen Y, Nicklas JA, Rainville IR, O’Neill JP. Development of long PCR techniques to analyze deletion mutations of the human hprt gene. Mutat Res. 1998;403:171–175. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

