High rates of infection with novel enterovirus variants in wild populations of mandrills and other old world monkey species
- PMID: 24623420
- PMCID: PMC4093852
- DOI: 10.1128/JVI.00088-14
High rates of infection with novel enterovirus variants in wild populations of mandrills and other old world monkey species
Abstract
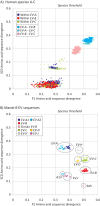
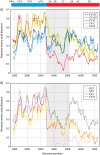
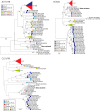
Enteroviruses (EVs) are a genetically and antigenically diverse group of viruses infecting humans. A mostly distinct set of EV variants have additionally been documented to infect wild apes and several, primarily captive, Old World monkey (OWM) species. To investigate the prevalence and genetic characteristics of EVs infecting OWMs in the wild, fecal samples from mandrills (Mandrillus sphinx) and other species collected in remote regions of southern Cameroon were screened for EV RNA. Remarkably high rates of EV positivity were detected in M. sphinx (100 of 102 screened), Cercocebus torquatus (7/7), and Cercopithecus cephus (2/4), with high viral loads indicative of active infection. Genetic characterization in VP4/VP2 and VP1 regions allowed EV variants to be assigned to simian species H (EV-H) and EV-J (including one or more new types), while seven matched simian EV-B variants, SA5 and EV110 (chimpanzee). Sequences from the remaining 70 formed a new genetic group distinct in VP4/2 and VP1 region from all currently recognized human or simian EV species. Complete genome sequences were obtained from three to determine their species assignment. In common with EV-J and the EV-A A13 isolate, new group sequences were chimeric, being most closely related to EV-A in capsid genes and to EV-B in the nonstructural gene region. Further recombination events created different groupings in 5' and 3' untranslated regions. While clearly a distinct EV group, the hybrid nature of new variants prevented their unambiguous classification as either members of a new species or as divergent members of EV-A using current International Committee on Taxonomy of Viruses (ICTV) assignment criteria.
Importance: This study is the first large-scale investigation of the frequency of infection and diversity of enteroviruses (EVs) infecting monkeys (primarily mandrills) in the wild. Our findings demonstrate extremely high frequencies of active infection (95%) among mandrills and other Old World monkey species inhabiting remote regions of Cameroon without human contact. EV variants detected were distinct from those infecting human populations, comprising members of enterovirus species B, J, and H and a large novel group of viruses most closely related to species A in the P1 region. The viral sequences obtained contribute substantially to our growing understanding of the genetic diversity of EVs and the existence of interspecies chimerism that characterizes the novel variants in the current study, as well as in previously characterized species A and J viruses infecting monkeys. The latter findings will contribute to future development of consensus criteria for species assignments in enteroviruses and other picornavirus genera.
Figures


References
-
- Knowles NJ, Hovi T, Hyypia T, King AMQ, Lindberg AM, Pallansch MA, Palmenberg AC, Simmonds P, Skern T, Stanway G, Yamashita T, Zell R. 2012. Picornaviridae, p 855–880 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA
-
- Heberling RL, Cheever FS. 1965. Some characteristics of the simian enteroviruses. Am. J. Epidemiol. 81:106–123 - PubMed
-
- Malherbe H, Harwin R. 1963. The cytopathic effects of vervet monkey viruses. S. Afr. Med. J. 37:407–411 - PubMed
-
- Hull RN, Minner JR, Mascoli CC. 1958. New viral agents recovered from tissue cultures of monkey kidney cells. III. Recovery of additional agents both from cultures of monkey tissues and directly from tissues and excreta. Am. J. Hyg. 68:31–44 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

