Emergence and evolution of avian H5N2 influenza viruses in chickens in Taiwan
- PMID: 24623422
- PMCID: PMC4019133
- DOI: 10.1128/JVI.00139-14
Emergence and evolution of avian H5N2 influenza viruses in chickens in Taiwan
Abstract
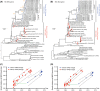
Sporadic activity by H5N2 influenza viruses has been observed in chickens in Taiwan from 2003 to 2012. The available information suggests that these viruses were generated by reassortment between a Mexican-like H5N2 virus and a local enzootic H6N1 virus. Yet the origin, prevalence, and pathogenicity of these H5N2 viruses have not been fully defined. Following the 2012 highly pathogenic avian influenza (HPAI) outbreaks, surveillance was conducted from December 2012 to July 2013 at a live-poultry wholesale market in Taipei. Our findings showed that H5N2 and H6N1 viruses cocirculated at low levels in chickens in Taiwan. Phylogenetic analyses revealed that all H5N2 viruses had hemagglutinin (HA) and neuraminidase (NA) genes derived from a 1994 Mexican-like virus, while their internal gene complexes were incorporated from the enzootic H6N1 virus lineage by multiple reassortment events. Pathogenicity studies demonstrated heterogeneous results even though all tested viruses had motifs (R-X-K/R-R) supportive of high pathogenicity. Serological surveys for common subtypes of avian viruses confirmed the prevalence of the H5N2 and H6N1 viruses in chickens and revealed an extraordinarily high seroconversion rate to an H9N2 virus, a subtype that is not found in Taiwan but is prevalent in mainland China. These findings suggest that reassortant H5N2 viruses, together with H6N1 viruses, have become established and enzootic in chickens throughout Taiwan and that a large-scale vaccination program might have been conducted locally that likely led to the introduction of the 1994 Mexican-like virus to Taiwan in 2003.
Importance: H5N2 avian influenza viruses first appeared in chickens in Taiwan in 2003 and caused a series of outbreaks afterwards. Phylogenetic analyses show that the chicken H5N2 viruses have H5 and N2 genes that are closely related to those of a vaccine strain originating from Mexico in 1994, while the contemporary duck H5N2 viruses in Taiwan belong to the Eurasian gene pool. The unusually high similarity of the chicken H5N2 viruses to the Mexican vaccine strain suggests that these viruses might have been introduced to Taiwan by using inadequately inactivated or attenuated vaccines. These chicken H5N2 viruses are developing varying levels of pathogenicity that could lead to significant consequences for the local poultry industry. These findings emphasize the need for strict quality control and competent oversight in the manufacture and usage of avian influenza virus vaccines and indicate that alternatives to widespread vaccination may be desirable.
Figures




References
-
- Banks J, Speidel ES, Moore E, Plowright L, Piccirillo A, Capua I, Cordioli P, Fioretti A, Alexander DJ. 2001. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch. Virol. 146:963–973. 10.1007/s007050170128 - DOI - PubMed
-
- Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F, Facchini M, Chiapponi C, Foni E, Cordioli P, Webby R, Barigazzi G, Webster RG, Donatelli I. 2004. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology 323:24–36. 10.1016/j.virol.2004.02.015 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

