MicroRNA-dependent regulation of transcription in non-small cell lung cancer
- PMID: 24625834
- PMCID: PMC3953115
- DOI: 10.1371/journal.pone.0090524
MicroRNA-dependent regulation of transcription in non-small cell lung cancer
Abstract
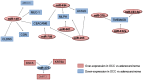
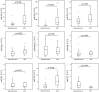
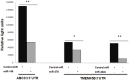
Squamous cell lung cancer (SCC) and adenocarcinoma are the most common histological subtypes of non-small cell lung cancer (NSCLC), and have been traditionally managed in the clinic as a single entity. Increasing evidence, however, illustrates the biological diversity of these two histological subgroups of lung cancer, and supports the need to improve our understanding of the molecular basis beyond the different phenotypes if we aim to develop more specific and individualized targeted therapy. The purpose of this study was to identify microRNA (miRNA)-dependent transcriptional regulation differences between SCC and adenocarcinoma histological lung cancer subtypes. In this work, paired miRNA (667 miRNAs by TaqMan Low Density Arrays (TLDA)) and mRNA profiling (Whole Genome 44 K array G112A, Agilent) was performed in tumor samples of 44 NSCLC patients. Nine miRNAs and 56 mRNAs were found to be differentially expressed in SCC versus adenocarcinoma samples. Eleven of these 56 mRNA were predicted as targets of the miRNAs identified to be differently expressed in these two histological conditions. Of them, 6 miRNAs (miR-149, miR-205, miR-375, miR-378, miR-422a and miR-708) and 9 target genes (CEACAM6, CGN, CLDN3, ABCC3, MLPH, ACSL5, TMEM45B, MUC1) were validated by quantitative PCR in an independent cohort of 41 lung cancer patients. Furthermore, the inverse correlation between mRNAs and microRNAs expression was also validated. These results suggest miRNA-dependent transcriptional regulation differences play an important role in determining key hallmarks of NSCLC, and may provide new biomarkers for personalized treatment strategies.
Conflict of interest statement
Figures





References
-
- Siegel R, Ward E, Brawley O, Jemal A (2011) Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 61: 212–236. - PubMed
-
- Minna JD, Roth JA, Gazdar AF (2002) Focus on lung cancer. Cancer Cell 1: 49–52. - PubMed
-
- Wistuba II (2007) Genetics of preneoplasia: lessons from lung cancer. Curr Mol Med 7: 3–14. - PubMed
-
- Petersen I, Bujard M, Petersen S, Wolf G, Goeze A, et al. (1997) Patterns of chromosomal imbalances in adenocarcinoma and squamous cell carcinoma of the lung. Cancer Res 57: 2331–2335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

