Clonality of HTLV-2 in natural infection
- PMID: 24626195
- PMCID: PMC3953477
- DOI: 10.1371/journal.ppat.1004006
Clonality of HTLV-2 in natural infection
Abstract
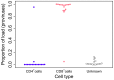
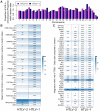
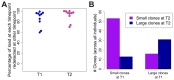
Human T-lymphotropic virus type 1 (HTLV-1) and type 2 (HTLV-2) both cause lifelong persistent infections, but differ in their clinical outcomes. HTLV-1 infection causes a chronic or acute T-lymphocytic malignancy in up to 5% of infected individuals whereas HTLV-2 has not been unequivocally linked to a T-cell malignancy. Virus-driven clonal proliferation of infected cells both in vitro and in vivo has been demonstrated in HTLV-1 infection. However, T-cell clonality in HTLV-2 infection has not been rigorously characterized. In this study we used a high-throughput approach in conjunction with flow cytometric sorting to identify and quantify HTLV-2-infected T-cell clones in 28 individuals with natural infection. We show that while genome-wide integration site preferences in vivo were similar to those found in HTLV-1 infection, expansion of HTLV-2-infected clones did not demonstrate the same significant association with the genomic environment of the integrated provirus. The proviral load in HTLV-2 is almost confined to CD8+ T-cells and is composed of a small number of often highly expanded clones. The HTLV-2 load correlated significantly with the degree of dispersion of the clone frequency distribution, which was highly stable over ∼8 years. These results suggest that there are significant differences in the selection forces that control the clonal expansion of virus-infected cells in HTLV-1 and HTLV-2 infection. In addition, our data demonstrate that strong virus-driven proliferation per se does not predispose to malignant transformation in oncoretroviral infections.
Conflict of interest statement
Niall Gormley is an employee of Illumina Inc, a public company that develops and markets systems for genetic analysis. This does not alter our adherence to all PLOS policies on sharing data and materials. The remaining authors have declared that no competing interests exist.
Figures


References
-
- Salemi M, Desmyter J, Vandamme AM (2000) Tempo and mode of human and simian T-lymphotropic virus (HTLV/STLV) evolution revealed by analyses of full-genome sequences. Mol Biol Evol 17: 374–386. - PubMed
-
- Roucoux DF, Murphy EL (2004) The epidemiology and disease outcomes of human T-lymphotropic virus type II. AIDS Rev 6: 144–154. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

