A study of the molecular mechanism of binding kinetics and long residence times of human CCR5 receptor small molecule allosteric ligands
- PMID: 24628038
- PMCID: PMC4105926
- DOI: 10.1111/bph.12683
A study of the molecular mechanism of binding kinetics and long residence times of human CCR5 receptor small molecule allosteric ligands
Abstract
Background and purpose: The human CCR5 receptor is a co-receptor for HIV-1 infection and a target for anti-viral therapy. A greater understanding of the binding kinetics of small molecule allosteric ligand interactions with CCR5 will lead to a better understanding of the binding process and may help discover new molecules that avoid resistance.
Experimental approach: Using [(3) H] maraviroc as a radioligand, a number of different binding protocols were employed in conjunction with simulations to determine rate constants, kinetic mechanism and mutant kinetic fingerprints for wild-type and mutant human CCR5 with maraviroc, aplaviroc and vicriviroc.
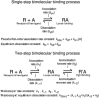
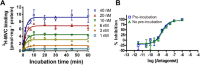
Key results: Kinetic characterization of maraviroc binding to the wild-type CCR5 was consistent with a two-step kinetic mechanism that involved an initial receptor-ligand complex (RA), which transitioned to a more stable complex, R'A, with at least a 13-fold increase in affinity. The dissociation rate from R'A, k-2 , was 1.2 × 10(-3) min(-1) . The maraviroc time-dependent transition was influenced by F85L, W86A, Y108A, I198A and Y251A mutations of CCR5.
Conclusions and implications: The interaction between maraviroc and CCR5 proceeded according to a multi-step kinetic mechanism, whereby initial mass action binding and later reorganizations of the initial maraviroc-receptor complex lead to a complex with longer residence time. Site-directed mutagenesis identified a kinetic fingerprint of residues that affected the binding kinetics, leading to the conclusion that allosteric ligand binding to CCR5 involved the rearrangement of the binding site in a manner specific to each allosteric ligand.
Keywords: CCR5; HIV-1 infection; allosteric; binding kinetics; maraviroc; molecular mechanism of action; pharmacology.
© 2014 The British Pharmacological Society.
Figures


References
-
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (IC50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. - PubMed
-
- Copeland R. The dynamics of drug-target interactions: drug-target residence time and its impact on efficacy and safety. Expert Opin Drug Discov. 2010;5:305–310. - PubMed
-
- Copeland RA. Conformational adaptation in drug-target interactions and residence time. Future Med Chem. 2011;3:1491–1501. - PubMed
-
- Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5:730–739. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

