Genomic convergence and network analysis approach to identify candidate genes in Alzheimer's disease
- PMID: 24628925
- PMCID: PMC4028079
- DOI: 10.1186/1471-2164-15-199
Genomic convergence and network analysis approach to identify candidate genes in Alzheimer's disease
Abstract
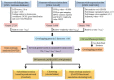
Background: Alzheimer's disease (AD) is one of the leading genetically complex and heterogeneous disorder that is influenced by both genetic and environmental factors. The underlying risk factors remain largely unclear for this heterogeneous disorder. In recent years, high throughput methodologies, such as genome-wide linkage analysis (GWL), genome-wide association (GWA) studies, and genome-wide expression profiling (GWE), have led to the identification of several candidate genes associated with AD. However, due to lack of consistency within their findings, an integrative approach is warranted. Here, we have designed a rank based gene prioritization approach involving convergent analysis of multi-dimensional data and protein-protein interaction (PPI) network modelling.
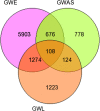
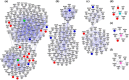
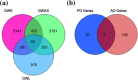
Results: Our approach employs integration of three different AD datasets- GWL,GWA and GWE to identify overlapping candidate genes ranked using a novel cumulative rank score (SR) based method followed by prioritization using clusters derived from PPI network. SR for each gene is calculated by addition of rank assigned to individual gene based on either p value or score in three datasets. This analysis yielded 108 plausible AD genes. Network modelling by creating PPI using proteins encoded by these genes and their direct interactors resulted in a layered network of 640 proteins. Clustering of these proteins further helped us in identifying 6 significant clusters with 7 proteins (EGFR, ACTB, CDC2, IRAK1, APOE, ABCA1 and AMPH) forming the central hub nodes. Functional annotation of 108 genes revealed their role in several biological activities such as neurogenesis, regulation of MAP kinase activity, response to calcium ion, endocytosis paralleling the AD specific attributes. Finally, 3 potential biochemical biomarkers were found from the overlap of 108 AD proteins with proteins from CSF and plasma proteome. EGFR and ACTB were found to be the two most significant AD risk genes.
Conclusions: With the assumption that common genetic signals obtained from different methodological platforms might serve as robust AD risk markers than candidates identified using single dimension approach, here we demonstrated an integrated genomic convergence approach for disease candidate gene prioritization from heterogeneous data sources linked to AD.
Figures



References
-
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–174. - PubMed
-
- Pedersen NL, Posner SF, Gatz M. Multiple-threshold models for genetic influences on age of onset for Alzheimer disease: findings in Swedish twins. Am J Med Genet. 2001;105(8):724–728. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

