A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity
- PMID: 24631838
- PMCID: PMC4012430
- DOI: 10.1158/2159-8290.CD-13-0929
A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity
Abstract
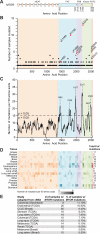
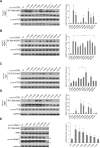
Genes encoding components of the PI3K-AKT-mTOR signaling axis are frequently mutated in cancer, but few mutations have been characterized in MTOR, the gene encoding the mTOR kinase. Using publicly available tumor genome sequencing data, we generated a comprehensive catalog of mTOR pathway mutations in cancer, identifying 33 MTOR mutations that confer pathway hyperactivation. The mutations cluster in six distinct regions in the C-terminal half of mTOR and occur in multiple cancer types, with one cluster particularly prominent in kidney cancer. The activating mutations do not affect mTOR complex assembly, but a subset reduces binding to the mTOR inhibitor DEPTOR. mTOR complex 1 (mTORC1) signaling in cells expressing various activating mutations remains sensitive to pharmacologic mTOR inhibition, but is partially resistant to nutrient deprivation. Finally, cancer cell lines with hyperactivating MTOR mutations display heightened sensitivity to rapamycin both in culture and in vivo xenografts, suggesting that such mutations confer mTOR pathway dependency.
Figures


References
-
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2(5):401–4. Epub 2012/05/17. doi: 2/5/401 [pii] 10.1158/2159-8290.CD-12-0095. PubMed PMID: 22588877. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

