T cell receptor bias for MHC: co-evolution or co-receptors?
- PMID: 24633202
- PMCID: PMC11113676
- DOI: 10.1007/s00018-014-1600-9
T cell receptor bias for MHC: co-evolution or co-receptors?
Abstract
In contrast to antibodies, which recognize antigens in native form, αβ T cell receptors (TCRs) only recognize antigens as peptide fragments bound to MHC molecules, a feature known as MHC restriction. The mechanism by which MHC restriction is imposed on the TCR repertoire is an unsolved problem that has generated considerable debate. Two principal models have been advanced to explain TCR bias for MHC. According to the germline model, MHC restriction is intrinsic to TCR structure because TCR and MHC molecules have co-evolved to conserve germline-encoded TCR sequences with the ability to bind MHC, while eliminating TCR sequences lacking MHC reactivity. According to the selection model, MHC restriction is not intrinsic to TCR structure, but is imposed by the CD4 and CD8 co-receptors that promote signaling by delivering the Src tyrosine kinase Lck to TCR-MHC complexes through co-receptor binding to MHC during positive selection. Here, we review the evidence for and against each model and conclude that both contribute to determining TCR specificity, although their relative contributions remain to be defined. Thus, TCR bias for MHC reflects not only germline-encoded TCR-MHC interactions but also the requirement to form a ternary complex with the CD4 or CD8 co-receptor that is geometrically competent to deliver a maturation signal to double-positive thymocytes during T cell selection.
Figures

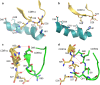
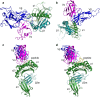

References
-
- Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, Chen W, Levin AM, Connolly JM, Zhu C, Kranz DM, Garcia KC. T cell receptor signaling is limited by docking geometry to peptide–major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

