The precarious prokaryotic chromosome
- PMID: 24633873
- PMCID: PMC4011006
- DOI: 10.1128/JB.00022-14
The precarious prokaryotic chromosome
Abstract
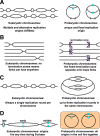
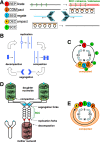
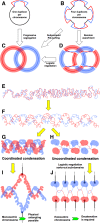
Evolutionary selection for optimal genome preservation, replication, and expression should yield similar chromosome organizations in any type of cells. And yet, the chromosome organization is surprisingly different between eukaryotes and prokaryotes. The nuclear versus cytoplasmic accommodation of genetic material accounts for the distinct eukaryotic and prokaryotic modes of genome evolution, but it falls short of explaining the differences in the chromosome organization. I propose that the two distinct ways to organize chromosomes are driven by the differences between the global-consecutive chromosome cycle of eukaryotes and the local-concurrent chromosome cycle of prokaryotes. Specifically, progressive chromosome segregation in prokaryotes demands a single duplicon per chromosome, while other "precarious" features of the prokaryotic chromosomes can be viewed as compensations for this severe restriction.
Figures


References
-
- Koonin EV. 2011. The logic of chance: the nature and origin of biological evolution. FT Press Science, Upper Saddle River, NJ
-
- Cavalier-Smith T. 1985. Eukaryote gene numbers, non-coding DNA and genome size, p 69–103 In Cavalier-Smith T. (ed), The evolution of genome size. John Wiley & Sons, Chichester, United Kingdom
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

