PolyTB: a genomic variation map for Mycobacterium tuberculosis
- PMID: 24637013
- PMCID: PMC4066953
- DOI: 10.1016/j.tube.2014.02.005
PolyTB: a genomic variation map for Mycobacterium tuberculosis
Abstract
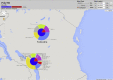
Tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb) is the second major cause of death from an infectious disease worldwide. Recent advances in DNA sequencing are leading to the ability to generate whole genome information in clinical isolates of M. tuberculosis complex (MTBC). The identification of informative genetic variants such as phylogenetic markers and those associated with drug resistance or virulence will help barcode Mtb in the context of epidemiological, diagnostic and clinical studies. Mtb genomic datasets are increasingly available as raw sequences, which are potentially difficult and computer intensive to process, and compare across studies. Here we have processed the raw sequence data (>1500 isolates, eight studies) to compile a catalogue of SNPs (n = 74,039, 63% non-synonymous, 51.1% in more than one isolate, i.e. non-private), small indels (n = 4810) and larger structural variants (n = 800). We have developed the PolyTB web-based tool (http://pathogenseq.lshtm.ac.uk/polytb) to visualise the resulting variation and important meta-data (e.g. in silico inferred strain-types, location) within geographical map and phylogenetic views. This resource will allow researchers to identify polymorphisms within candidate genes of interest, as well as examine the genomic diversity and distribution of strains. PolyTB source code is freely available to researchers wishing to develop similar tools for their pathogen of interest.
Keywords: Database; Genomics; Molecular epidemiology; Mycobacterium tuberculosis; Software; Whole-genome sequencing.
Copyright © 2014 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
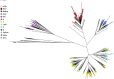



Similar articles
-
PhyTB: Phylogenetic tree visualisation and sample positioning for M. tuberculosis.BMC Bioinformatics. 2015 May 13;16(1):155. doi: 10.1186/s12859-015-0603-3. BMC Bioinformatics. 2015. PMID: 25968323 Free PMC article.
-
[Future prospects of molecular epidemiology in tuberculosis].Kekkaku. 2009 Dec;84(12):783-4. Kekkaku. 2009. PMID: 20077862 Japanese.
-
Whole Genome Sequencing of Mycobacterium tuberculosis Isolates From Extrapulmonary Sites.OMICS. 2017 Jul;21(7):413-425. doi: 10.1089/omi.2017.0070. OMICS. 2017. PMID: 28692415 Free PMC article.
-
Bioinformatics tools and databases for whole genome sequence analysis of Mycobacterium tuberculosis.Infect Genet Evol. 2016 Nov;45:359-368. doi: 10.1016/j.meegid.2016.09.013. Epub 2016 Sep 13. Infect Genet Evol. 2016. PMID: 27637931 Review.
-
Large genomics datasets shed light on the evolution of the Mycobacterium tuberculosis complex.Infect Genet Evol. 2019 Aug;72:10-15. doi: 10.1016/j.meegid.2019.02.028. Epub 2019 Feb 26. Infect Genet Evol. 2019. PMID: 30822550 Review.
Cited by
-
Exploiting Homoplasy in Genome-Wide Association Studies to Enhance Identification of Antibiotic-Resistance Mutations in Bacterial Genomes.Evol Bioinform Online. 2020 Jul 27;16:1176934320944932. doi: 10.1177/1176934320944932. eCollection 2020. Evol Bioinform Online. 2020. PMID: 32782426 Free PMC article.
-
A robust SNP barcode for typing Mycobacterium tuberculosis complex strains.Nat Commun. 2014 Sep 1;5:4812. doi: 10.1038/ncomms5812. Nat Commun. 2014. PMID: 25176035 Free PMC article.
-
A Rapid Drug Resistance Genotyping Workflow for Mycobacterium tuberculosis, Using Targeted Isothermal Amplification and Nanopore Sequencing.Microbiol Spectr. 2021 Dec 22;9(3):e0061021. doi: 10.1128/Spectrum.00610-21. Epub 2021 Nov 24. Microbiol Spectr. 2021. PMID: 34817282 Free PMC article.
-
Diversity of Mycobacterium tuberculosis in the Middle Fly District of Western Province, Papua New Guinea: microbead-based spoligotyping using DNA from Ziehl-Neelsen-stained microscopy preparations.Sci Rep. 2019 Oct 29;9(1):15549. doi: 10.1038/s41598-019-51892-5. Sci Rep. 2019. PMID: 31664101 Free PMC article.
-
Spoligotyping analysis of Mycobacterium tuberculosis in Khyber Pakhtunkhwa area, Pakistan.Infect Drug Resist. 2019 May 20;12:1363-1369. doi: 10.2147/IDR.S198314. eCollection 2019. Infect Drug Resist. 2019. PMID: 31190924 Free PMC article.
References
-
- Abubakar I., Zignol M., Falzon D., Raviglione M., Ditiu L., Masham S., Adetifa I., Ford N., Cox H., Lawn S.D., Marais B.J., McHugh T.D., Mwaba P., Bates M., Lipman M., Zijenah L., Logan S., McNerney R., Zumla A., Sarda K., Nahid P., Hoelscher M., Pletschette M., Memish Z.a., Kim P., Hafner R., Cole S., Migliori G.B., Maeurer M., Schito M., Zumla A. Drug-resistant tuberculosis: time for visionary political leadership. Lancet Infect Dis. 2013;13:529–539. - PubMed
-
- Garcia-Betancur J.C., Menendez M.C., Del Portillo P., Garcia M.J. Alignment of multiple complete genomes suggests that gene rearrangements may contribute towards the speciation of Mycobacteria. Infect Genet Evol. 2011;12:819–826. - PubMed
-
- Blouin Y., Hauck Y., Soler C., Fabre M., Vong R., Dehan C., Cazajous G., Massoure P.-L., Kraemer P., Jenkins A., Garnotel E., Pourcel C., Vergnaud G. Significance of the identification in the horn of Africa of an exceptionally deep branching Mycobacterium tuberculosis clade. PloS One. 2012;7:e52841. - PMC - PubMed
-
- Comas I., Coscolla M., Luo T., Borrell S., Holt K.E., Kato-Maeda M., Parkhill J., Malla B., Berg S., Thwaites G., Yeboah-Manu D., Bothamley G., Mei J., Wei L., Bentley S., Harris S.R., Niemann S., Diel R., Aseffa A., Gao Q., Young D., Gagneux S. Out-of-Africa migration and neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. - PMC - PubMed
-
- Supply P., Marceau M., Mangenot S., Roche D., Rouanet C., Khanna V., Majlessi L., Criscuolo A., Tap J., Pawlik A., Fiette L., Orgeur M., Fabre M., Parmentier C., Frigui W., Simeone R., Boritsch E.C., Debrie A.-S., Willery E., Walker D., Quail M.a., Ma L., Bouchier C., Salvignol G., Sayes F., Cascioferro A., Seemann T., Barbe V., Locht C., Gutierrez M.-C., Leclerc C., Bentley S.D., Stinear T.P., Brisse S., Médigue C., Parkhill J., Cruveiller S., Brosch R. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 2013;45:172–179. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

