DNA methylation is required for the control of stem cell differentiation in the small intestine
- PMID: 24637118
- PMCID: PMC3967052
- DOI: 10.1101/gad.230318.113
DNA methylation is required for the control of stem cell differentiation in the small intestine
Abstract
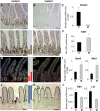
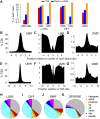
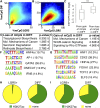
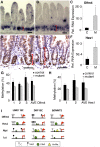
The mammalian intestinal epithelium has a unique organization in which crypts harboring stem cells produce progenitors and finally clonal populations of differentiated cells. Remarkably, the epithelium is replaced every 3-5 d throughout adult life. Disrupted maintenance of the intricate balance of proliferation and differentiation leads to loss of epithelial integrity or barrier function or to cancer. There is a tight correlation between the epigenetic status of genes and expression changes during differentiation; however, the mechanism of how changes in DNA methylation direct gene expression and the progression from stem cells to their differentiated descendants is unclear. Using conditional gene ablation of the maintenance methyltransferase Dnmt1, we demonstrate that reducing DNA methylation causes intestinal crypt expansion in vivo. Determination of the base-resolution DNA methylome in intestinal stem cells and their differentiated descendants shows that DNA methylation is dynamic at enhancers, which are often associated with genes important for both stem cell maintenance and differentiation. We establish that the loss of DNA methylation at intestinal stem cell gene enhancers causes inappropriate gene expression and delayed differentiation.
Keywords: DNA methylation; Dnmt1; intestinal stem cell.
Figures



References
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 - PubMed
-
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457: 608–611 - PubMed
-
- Benoit YD, Pare F, Francoeur C, Jean D, Tremblay E, Boudreau F, Escaffit F, Beaulieu JF 2010. Cooperation between HNF-1α, Cdx2, and GATA-4 in initiating an enterocytic differentiation program in a normal human intestinal epithelial progenitor cell line. Am J Physiol Gastrointest Liver Physiol 298: G504–G517 - PMC - PubMed
-
- Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M 2008. Computational analysis of small RNA cloning data. Methods 44: 13–21 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
