BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups
- PMID: 24638167
- PMCID: PMC3978272
- DOI: 10.1084/jem.20130977
BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups
Erratum in
-
BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups.J Exp Med. 2015 Feb 9;212(2):281. doi: 10.1084/jem.2013097701202015c. Epub 2015 Feb 2. J Exp Med. 2015. PMID: 25646268 Free PMC article. No abstract available.
Abstract
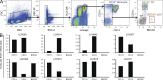
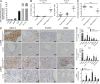
Langerhans cell histiocytosis (LCH) is a clonal disorder with elusive etiology, characterized by the accumulation of CD207(+) dendritic cells (DCs) in inflammatory lesions. Recurrent BRAF-V600E mutations have been reported in LCH. In this study, lesions from 100 patients were genotyped, and 64% carried the BRAF-V600E mutation within infiltrating CD207(+) DCs. BRAF-V600E expression in tissue DCs did not define specific clinical risk groups but was associated with increased risk of recurrence. Strikingly, we found that patients with active, high-risk LCH also carried BRAF-V600E in circulating CD11c(+) and CD14(+) fractions and in bone marrow (BM) CD34(+) hematopoietic cell progenitors, whereas the mutation was restricted to lesional CD207(+) DC in low-risk LCH patients. Importantly, BRAF-V600E expression in DCs was sufficient to drive LCH-like disease in mice. Consistent with our findings in humans, expression of BRAF-V600E in BM DC progenitors recapitulated many features of the human high-risk LCH, whereas BRAF-V600E expression in differentiated DCs more closely resembled low-risk LCH. We therefore propose classification of LCH as a myeloid neoplasia and hypothesize that high-risk LCH arises from somatic mutation of a hematopoietic progenitor, whereas low-risk disease arises from somatic mutation of tissue-restricted precursor DCs.
Figures





References
-
- Allen C.E., Li L., Peters T.L., Leung H.C., Yu A., Man T.K., Gurusiddappa S., Phillips M.T., Hicks M.J., Gaikwad A., et al. 2010. Cell-specific gene expression in Langerhans cell histiocytosis lesions reveals a distinct profile compared with epidermal Langerhans cells. J. Immunol. 184:4557–4567 10.4049/jimmunol.0902336 - DOI - PMC - PubMed
-
- Bernard F., Thomas C., Bertrand Y., Munzer M., Landman Parker J., Ouache M., Colin V.M., Perel Y., Chastagner P., Vermylen C., Donadieu J. 2005. Multi-centre pilot study of 2-chlorodeoxyadenosine and cytosine arabinoside combined chemotherapy in refractory Langerhans cell histiocytosis with haematological dysfunction. Eur. J. Cancer. 41:2682–2689 10.1016/j.ejca.2005.02.007 - DOI - PubMed
-
- Birbeck M.S.C., Breathnach A.S., Everall J.D. 1961. An electron microscope study of basal melanocytes and high-level clear cells (Langerhans cell) in vitiligo. J. Invest. Dermatol. 37:51–64 10.1038/jid.1961.7 - DOI
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA154489/CA/NCI NIH HHS/United States
- R01 CA154947/CA/NCI NIH HHS/United States
- R01 CA161373/CA/NCI NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- R01 CA154947A/CA/NCI NIH HHS/United States
- R01 AI089987/AI/NIAID NIH HHS/United States
- K12 CA090433/CA/NCI NIH HHS/United States
- U01 AI095611/AI/NIAID NIH HHS/United States
- R01 AI10008/AI/NIAID NIH HHS/United States
- T32 GM088129/GM/NIGMS NIH HHS/United States
- U19 AI089987/AI/NIAID NIH HHS/United States
- P50 CA126752/CA/NCI NIH HHS/United States
- P50CA126752/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

